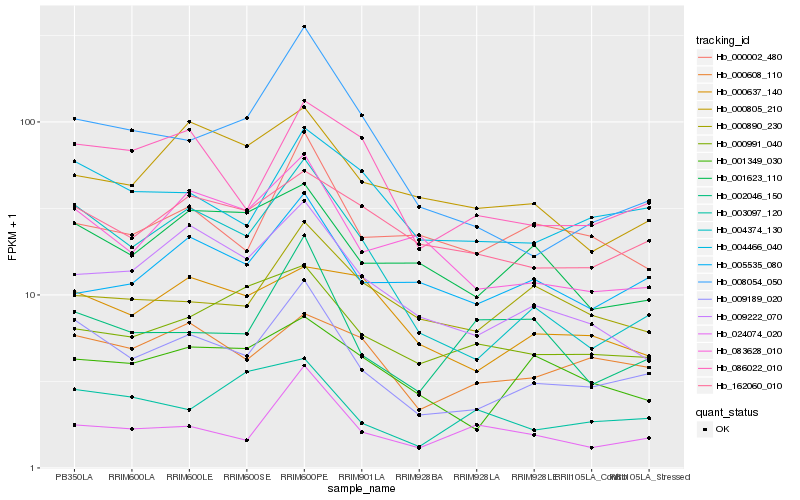
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009189_020 |
0.0 |
- |
- |
PREDICTED: CASP-like protein 4B4 [Jatropha curcas] |
| 2 |
Hb_004374_130 |
0.1200407371 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA1B-like [Jatropha curcas] |
| 3 |
Hb_004466_040 |
0.1332851555 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 4 |
Hb_083628_010 |
0.1425527845 |
- |
- |
PREDICTED: probable serine protease EDA2 [Jatropha curcas] |
| 5 |
Hb_086022_010 |
0.1428373204 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 6 |
Hb_009222_070 |
0.1497647613 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 7 |
Hb_000608_110 |
0.1517047376 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 8 |
Hb_001623_110 |
0.1562290016 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_162060_010 |
0.1590501272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001349_030 |
0.1619757191 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000002_480 |
0.1631016033 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_024074_020 |
0.164483877 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 13 |
Hb_000991_040 |
0.166004722 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 14 |
Hb_002046_150 |
0.1675406223 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g15080 [Jatropha curcas] |
| 15 |
Hb_000805_210 |
0.1679368182 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 16 |
Hb_003097_120 |
0.1689068108 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 17 |
Hb_008054_050 |
0.1694279218 |
- |
- |
PREDICTED: uncharacterized protein LOC101212042 [Cucumis sativus] |
| 18 |
Hb_000890_230 |
0.1694396623 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 19 |
Hb_005535_080 |
0.1696579562 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 20 |
Hb_000637_140 |
0.1717341879 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |