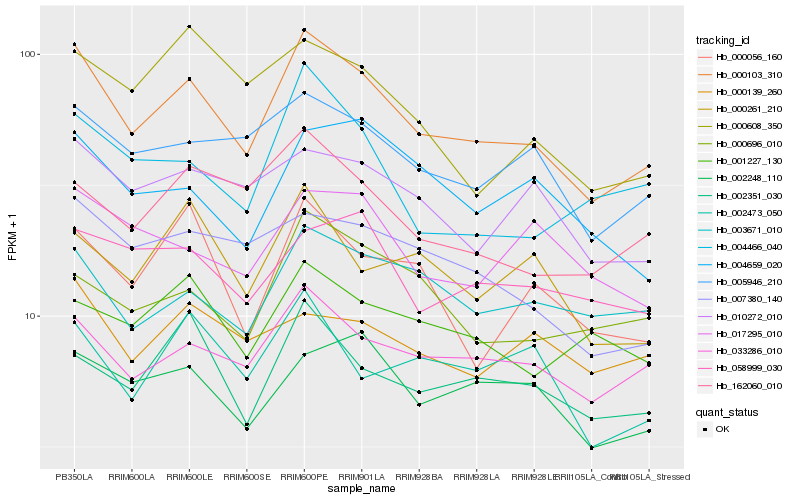
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_310 |
0.0 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |
| 2 |
Hb_033286_010 |
0.0860988944 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 3 |
Hb_005946_210 |
0.0951250583 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 4 |
Hb_003671_010 |
0.1009169484 |
- |
- |
PREDICTED: protein transport protein SFT2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002351_030 |
0.1017899586 |
- |
- |
catalytic, putative [Ricinus communis] |
| 6 |
Hb_162060_010 |
0.1023595413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002248_110 |
0.1032931817 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 8 |
Hb_010272_010 |
0.1035119619 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 9 |
Hb_007380_140 |
0.1035597491 |
- |
- |
PREDICTED: probable galacturonosyltransferase 4 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000696_010 |
0.1061958659 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 11 |
Hb_000056_160 |
0.1081788641 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP23 isoform X1 [Jatropha curcas] |
| 12 |
Hb_058999_030 |
0.1088047259 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_004466_040 |
0.1089047924 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 14 |
Hb_000261_210 |
0.1098422929 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 15 |
Hb_001227_130 |
0.1104240923 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 16 |
Hb_004659_020 |
0.1110808135 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 17 |
Hb_002473_050 |
0.1120353383 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_017295_010 |
0.1137128239 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 19 |
Hb_000139_260 |
0.1151712039 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 20 |
Hb_000608_350 |
0.1163827017 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |