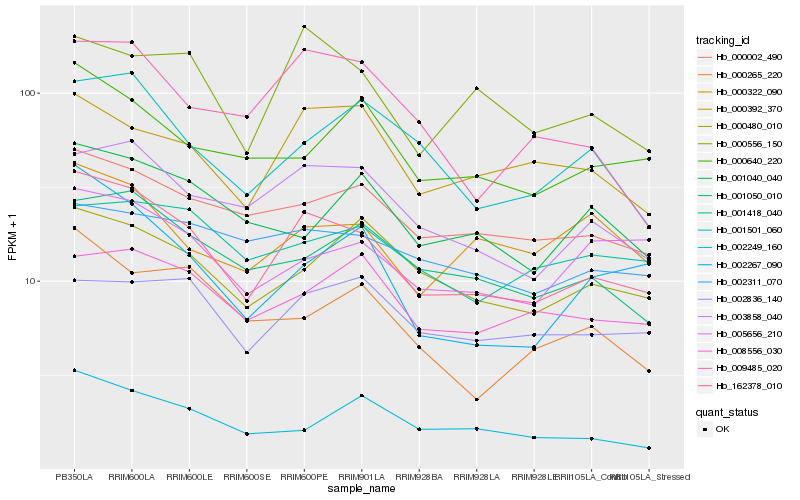
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162378_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_20480 [Jatropha curcas] |
| 2 |
Hb_003858_040 |
0.1108235691 |
- |
- |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 3 |
Hb_000002_490 |
0.1178319254 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 4 |
Hb_001050_010 |
0.1225345246 |
- |
- |
Advillin [Gossypium arboreum] |
| 5 |
Hb_000556_150 |
0.1249109488 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 6 |
Hb_000480_010 |
0.1295078382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000392_370 |
0.130532313 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 8 |
Hb_001501_060 |
0.131271406 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002267_090 |
0.1333657611 |
- |
- |
Cytochrome c oxidase assembly protein COX19, putative [Ricinus communis] |
| 10 |
Hb_002249_160 |
0.1338932788 |
- |
- |
- |
| 11 |
Hb_008556_030 |
0.1339258582 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005656_210 |
0.134802844 |
- |
- |
PREDICTED: uncharacterized protein LOC105637440 [Jatropha curcas] |
| 13 |
Hb_000640_220 |
0.1353554693 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 14 |
Hb_009485_020 |
0.1363493945 |
- |
- |
PREDICTED: membrane steroid-binding protein 2-like [Jatropha curcas] |
| 15 |
Hb_000322_090 |
0.1395682649 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_002836_140 |
0.1395720545 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 17 |
Hb_000265_220 |
0.1395764713 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 18 |
Hb_001040_040 |
0.1414593888 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 19 |
Hb_002311_070 |
0.1417805548 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 20 |
Hb_001418_040 |
0.1420267677 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |