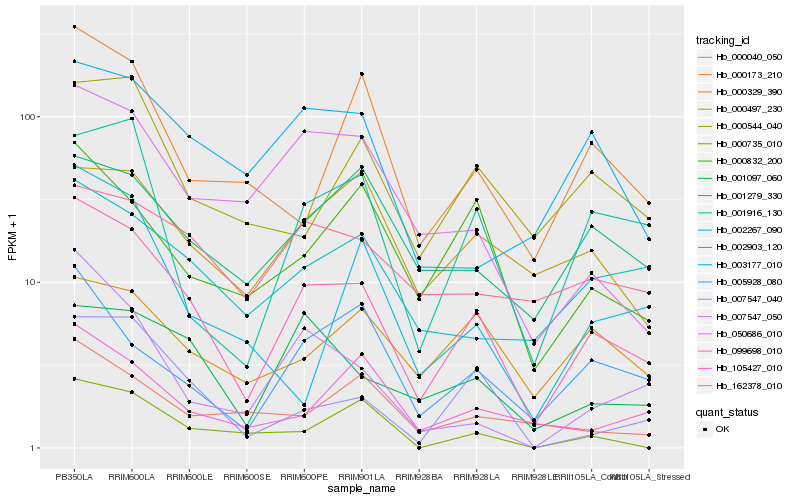
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_105427_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_007547_040 |
0.1856649517 |
- |
- |
Auxin response factor 9 [Aegilops tauschii] |
| 3 |
Hb_050686_010 |
0.1952727053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001097_060 |
0.1953717593 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 8 [Jatropha curcas] |
| 5 |
Hb_002267_090 |
0.2010709532 |
- |
- |
Cytochrome c oxidase assembly protein COX19, putative [Ricinus communis] |
| 6 |
Hb_005928_080 |
0.2029764133 |
- |
- |
PREDICTED: probable galacturonosyltransferase 6 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001916_130 |
0.2038863913 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |
| 8 |
Hb_000832_200 |
0.204311795 |
- |
- |
2-oxoglutarate dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_007547_050 |
0.2044828703 |
- |
- |
Auxin response factor 9 [Aegilops tauschii] |
| 10 |
Hb_000544_040 |
0.2047472039 |
- |
- |
PREDICTED: mitogen-activated protein kinase 9-like [Jatropha curcas] |
| 11 |
Hb_099698_010 |
0.2059208933 |
- |
- |
- |
| 12 |
Hb_000173_210 |
0.2071184944 |
- |
- |
PREDICTED: serine incorporator 3 [Jatropha curcas] |
| 13 |
Hb_003177_010 |
0.2108047338 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 14 |
Hb_162378_010 |
0.2168670249 |
- |
- |
hypothetical protein JCGZ_20480 [Jatropha curcas] |
| 15 |
Hb_000040_050 |
0.2172197119 |
- |
- |
PREDICTED: uncharacterized protein LOC102617857 isoform X1 [Citrus sinensis] |
| 16 |
Hb_000735_010 |
0.2178888912 |
- |
- |
- |
| 17 |
Hb_001279_330 |
0.2183855678 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 18 |
Hb_000497_230 |
0.2210196416 |
- |
- |
PREDICTED: probable protein S-acyltransferase 6 [Jatropha curcas] |
| 19 |
Hb_000329_390 |
0.2237364221 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002903_120 |
0.2244488417 |
- |
- |
- |