| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
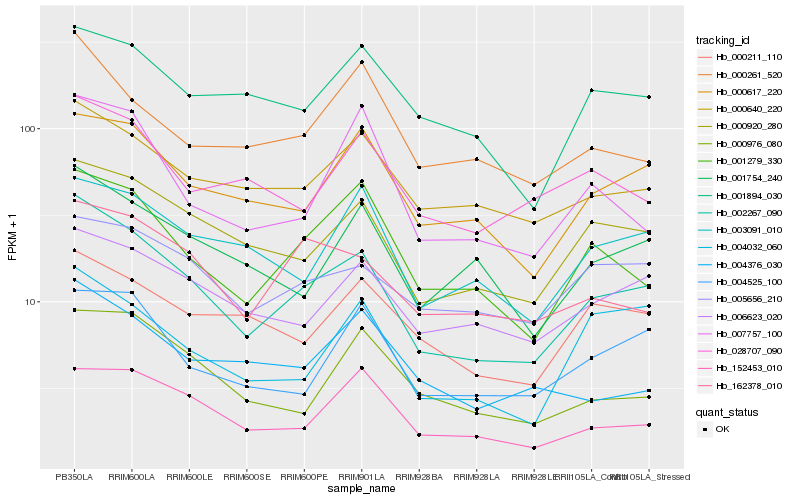
Hb_002267_090 |
0.0 |
- |
- |
Cytochrome c oxidase assembly protein COX19, putative [Ricinus communis] |
| 2 |
Hb_000920_280 |
0.104938886 |
- |
- |
Protein SENSITIVITY TO RED LIGHT REDUCED, putative [Ricinus communis] |
| 3 |
Hb_001279_330 |
0.1234090736 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 4 |
Hb_000617_220 |
0.1254367673 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 5 |
Hb_000640_220 |
0.1282206824 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 6 |
Hb_006623_020 |
0.1289033435 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001754_240 |
0.1297773349 |
- |
- |
PREDICTED: mitochondrial thiamine pyrophosphate carrier isoform X1 [Jatropha curcas] |
| 8 |
Hb_162378_010 |
0.1333657611 |
- |
- |
hypothetical protein JCGZ_20480 [Jatropha curcas] |
| 9 |
Hb_004525_100 |
0.1359591467 |
- |
- |
hypothetical protein JCGZ_18418 [Jatropha curcas] |
| 10 |
Hb_005656_210 |
0.1368627295 |
- |
- |
PREDICTED: uncharacterized protein LOC105637440 [Jatropha curcas] |
| 11 |
Hb_152453_010 |
0.1377153965 |
- |
- |
PREDICTED: putative transferase CAF17 homolog, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000211_110 |
0.1395784876 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 35 kDa protein isoform X1 [Jatropha curcas] |
| 13 |
Hb_003091_010 |
0.1405248349 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 14 |
Hb_004032_060 |
0.1421253757 |
- |
- |
elongation factor 1 gamma-like protein, partial [Ipomoea nil] |
| 15 |
Hb_001894_030 |
0.1423186926 |
- |
- |
Nucleic acid-binding, OB-fold-like protein [Theobroma cacao] |
| 16 |
Hb_000976_080 |
0.1423309441 |
- |
- |
hypothetical protein VITISV_034376 [Vitis vinifera] |
| 17 |
Hb_004376_030 |
0.1425886645 |
transcription factor |
TF Family: mTERF |
hypothetical protein PHAVU_005G060900g [Phaseolus vulgaris] |
| 18 |
Hb_000261_520 |
0.1431368153 |
- |
- |
PREDICTED: uncharacterized protein LOC105633192 [Jatropha curcas] |
| 19 |
Hb_028707_090 |
0.1438449365 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 20 |
Hb_007757_100 |
0.1441524119 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |