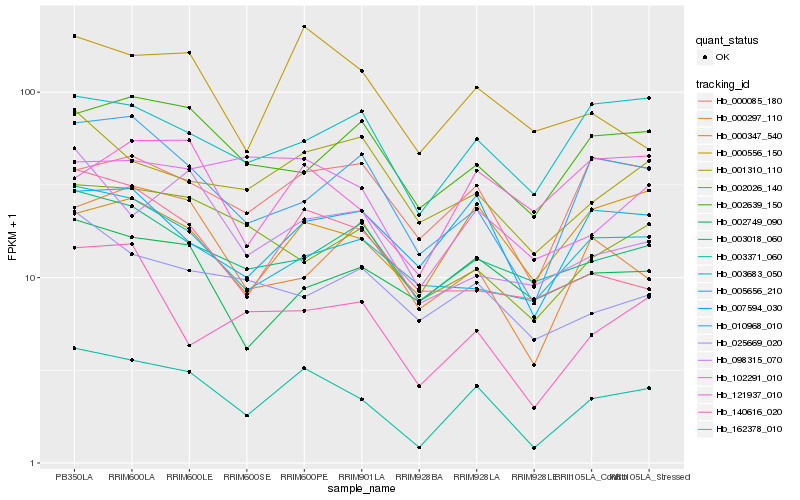
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003371_060 |
0.0 |
- |
- |
PREDICTED: cyclic pyranopterin monophosphate synthase, mitochondrial isoform X3 [Populus euphratica] |
| 2 |
Hb_010968_010 |
0.1585034727 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 3 |
Hb_000556_150 |
0.1674011019 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 4 |
Hb_007594_030 |
0.1692044534 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 5 |
Hb_002026_140 |
0.1704665799 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 6 |
Hb_162378_010 |
0.1706344205 |
- |
- |
hypothetical protein JCGZ_20480 [Jatropha curcas] |
| 7 |
Hb_005656_210 |
0.1708120443 |
- |
- |
PREDICTED: uncharacterized protein LOC105637440 [Jatropha curcas] |
| 8 |
Hb_140616_020 |
0.171367436 |
- |
- |
- |
| 9 |
Hb_001310_110 |
0.1738611237 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_000085_180 |
0.1740202866 |
- |
- |
PREDICTED: syntaxin-52-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_003018_060 |
0.1740611917 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 12 |
Hb_102291_010 |
0.1743464704 |
- |
- |
PREDICTED: histone chaperone ASF1B [Populus euphratica] |
| 13 |
Hb_098315_070 |
0.1756949421 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105636030 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002639_150 |
0.1780503476 |
- |
- |
- |
| 15 |
Hb_025669_020 |
0.1780628218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000297_110 |
0.1795497223 |
- |
- |
PREDICTED: uncharacterized protein LOC105634718 [Jatropha curcas] |
| 17 |
Hb_003683_050 |
0.1800553311 |
- |
- |
PREDICTED: uncharacterized protein LOC105650619 [Jatropha curcas] |
| 18 |
Hb_000347_540 |
0.1829204409 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_121937_010 |
0.1829388642 |
- |
- |
hypothetical protein B456_013G261900 [Gossypium raimondii] |
| 20 |
Hb_002749_090 |
0.1840732301 |
- |
- |
- |