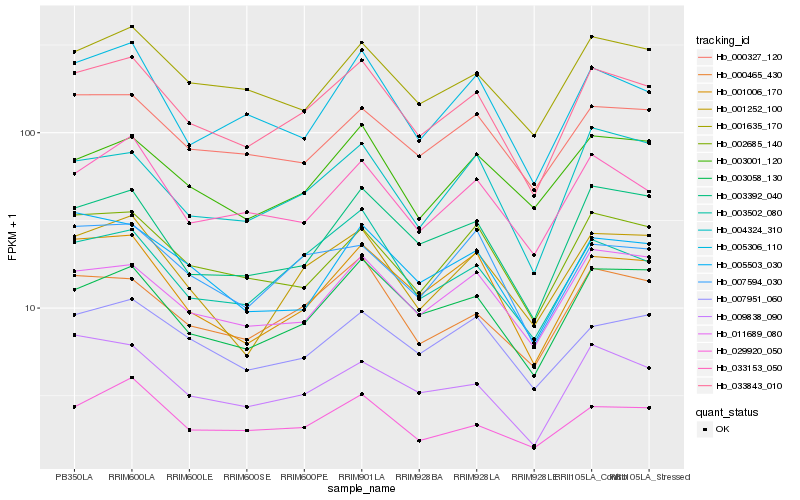
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033843_010 |
0.0 |
- |
- |
PREDICTED: cell division control protein 2 homolog A isoform X1 [Jatropha curcas] |
| 2 |
Hb_000465_430 |
0.0633017255 |
- |
- |
PREDICTED: uncharacterized protein LOC105632325 [Jatropha curcas] |
| 3 |
Hb_003502_080 |
0.0647074242 |
- |
- |
vacuolar protein sorting, putative [Ricinus communis] |
| 4 |
Hb_003058_130 |
0.0651110295 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A isoform X1 [Jatropha curcas] |
| 5 |
Hb_001006_170 |
0.0716867533 |
- |
- |
Endonuclease V, putative [Ricinus communis] |
| 6 |
Hb_033153_050 |
0.0727450559 |
transcription factor |
TF Family: VOZ |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003392_040 |
0.0728881107 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Jatropha curcas] |
| 8 |
Hb_001252_100 |
0.0732396051 |
- |
- |
PREDICTED: CASP-like protein 5B2 [Jatropha curcas] |
| 9 |
Hb_001635_170 |
0.0739417188 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 10 |
Hb_002685_140 |
0.0748349404 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 11 |
Hb_000327_120 |
0.0755120621 |
- |
- |
FGFR1 oncogene partner, putative [Ricinus communis] |
| 12 |
Hb_029920_050 |
0.0766621435 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 13 |
Hb_007951_060 |
0.0766745999 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 14 |
Hb_011689_080 |
0.0772116157 |
- |
- |
PREDICTED: uncharacterized protein LOC105631867 [Jatropha curcas] |
| 15 |
Hb_005306_110 |
0.0786258722 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 16 |
Hb_007594_030 |
0.0797067247 |
- |
- |
PREDICTED: cucumber peeling cupredoxin-like [Jatropha curcas] |
| 17 |
Hb_009838_090 |
0.0804887165 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004324_310 |
0.081169694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003001_120 |
0.0825373239 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005503_030 |
0.0827234313 |
- |
- |
PREDICTED: protein SCO1 homolog 1, mitochondrial [Jatropha curcas] |