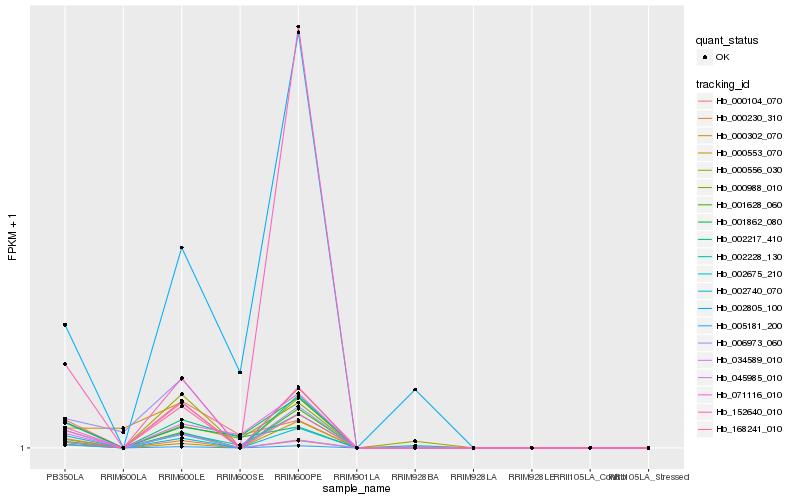
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002740_070 |
0.0 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 2 |
Hb_000230_310 |
0.1525182466 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 3 |
Hb_071116_010 |
0.1567645818 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 4 |
Hb_000553_070 |
0.167612015 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001862_080 |
0.2371103514 |
- |
- |
hypothetical protein RCOM_1154950 [Ricinus communis] |
| 6 |
Hb_034589_010 |
0.2501602287 |
- |
- |
hypothetical protein MIMGU_mgv1a020753mg, partial [Erythranthe guttata] |
| 7 |
Hb_002228_130 |
0.2678461695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001628_060 |
0.2729298181 |
- |
- |
PREDICTED: beta-galactosidase 13-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_002675_210 |
0.2755185333 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 10 |
Hb_045985_010 |
0.2870894171 |
- |
- |
hypothetical protein JCGZ_00291 [Jatropha curcas] |
| 11 |
Hb_152640_010 |
0.2942661426 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 12 |
Hb_002805_100 |
0.3001772784 |
- |
- |
Receptor-like protein 12 [Morus notabilis] |
| 13 |
Hb_000302_070 |
0.3063896429 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 14 |
Hb_168241_010 |
0.3066412162 |
- |
- |
hypothetical protein POPTR_0013s14620g, partial [Populus trichocarpa] |
| 15 |
Hb_002217_410 |
0.3091615793 |
- |
- |
hypothetical protein L484_027883 [Morus notabilis] |
| 16 |
Hb_000104_070 |
0.3221317592 |
- |
- |
PREDICTED: citrate-binding protein-like [Jatropha curcas] |
| 17 |
Hb_005181_200 |
0.3223030838 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein RGL1-like [Jatropha curcas] |
| 18 |
Hb_006973_060 |
0.3274650602 |
- |
- |
Uncharacterized protein TCM_038475 [Theobroma cacao] |
| 19 |
Hb_000556_030 |
0.3283472488 |
- |
- |
PREDICTED: probable carboxylesterase 9 [Jatropha curcas] |
| 20 |
Hb_000988_010 |
0.3361685885 |
- |
- |
hypothetical protein POPTR_0004s17730g [Populus trichocarpa] |