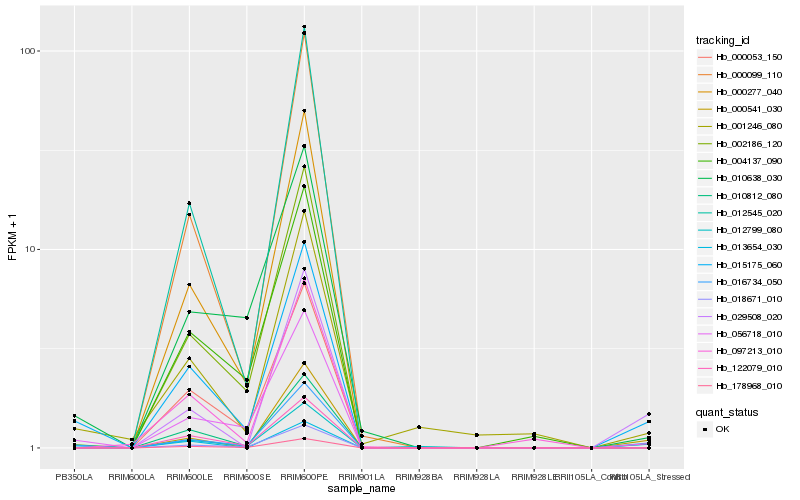
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015175_060 |
0.0 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 2 |
Hb_016734_050 |
0.0781814146 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 3 |
Hb_056718_010 |
0.1577115816 |
- |
- |
hypothetical protein JCGZ_11777 [Jatropha curcas] |
| 4 |
Hb_013654_030 |
0.1658686551 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 5 |
Hb_012799_080 |
0.1671668273 |
transcription factor |
TF Family: MYB |
Myb domain protein 18, putative [Theobroma cacao] |
| 6 |
Hb_001246_080 |
0.1683711771 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 7 |
Hb_010812_080 |
0.1750778459 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 8 |
Hb_122079_010 |
0.1756089343 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 9 |
Hb_000053_150 |
0.1759558804 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 10 |
Hb_000277_040 |
0.1777207675 |
- |
- |
- |
| 11 |
Hb_178968_010 |
0.1811116527 |
- |
- |
hypothetical protein JCGZ_15830 [Jatropha curcas] |
| 12 |
Hb_097213_010 |
0.1820178967 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Populus euphratica] |
| 13 |
Hb_012545_020 |
0.1822569851 |
- |
- |
PREDICTED: uncharacterized protein LOC105633903 [Jatropha curcas] |
| 14 |
Hb_002186_120 |
0.1828130652 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X5 [Populus euphratica] |
| 15 |
Hb_010638_030 |
0.1828708071 |
- |
- |
oleosin1 [Plukenetia volubilis] |
| 16 |
Hb_018671_010 |
0.1835637599 |
- |
- |
- |
| 17 |
Hb_000099_110 |
0.1845773696 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 18 |
Hb_004137_090 |
0.1847731115 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 19 |
Hb_029508_020 |
0.1867862942 |
- |
- |
- |
| 20 |
Hb_000541_030 |
0.1877239886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |