| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
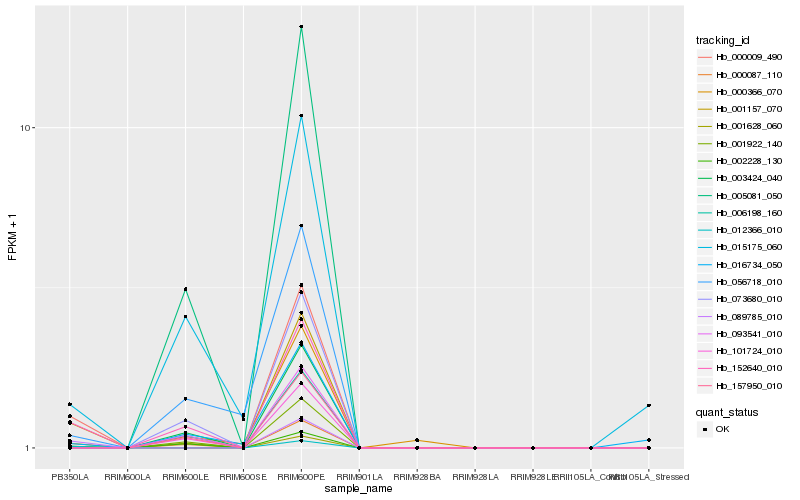
Hb_152640_010 |
0.0 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Populus euphratica] |
| 2 |
Hb_002228_130 |
0.1240028322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001628_060 |
0.2063708101 |
- |
- |
PREDICTED: beta-galactosidase 13-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_015175_060 |
0.2074550616 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 5 |
Hb_001157_070 |
0.2129684633 |
- |
- |
- |
| 6 |
Hb_000009_490 |
0.2133401897 |
- |
- |
hypothetical protein POPTR_0007s12040g [Populus trichocarpa] |
| 7 |
Hb_089785_010 |
0.2239369696 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Fragaria vesca subsp. vesca] |
| 8 |
Hb_012366_010 |
0.2265666308 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 9 |
Hb_056718_010 |
0.2281644548 |
- |
- |
hypothetical protein JCGZ_11777 [Jatropha curcas] |
| 10 |
Hb_016734_050 |
0.2295850512 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000366_070 |
0.2303470299 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.2 [Jatropha curcas] |
| 12 |
Hb_005081_050 |
0.2361386015 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Prunus mume] |
| 13 |
Hb_073680_010 |
0.2361485868 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_157950_010 |
0.2361496387 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003424_040 |
0.2361653383 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 16 |
Hb_001922_140 |
0.2361906281 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000087_110 |
0.236220094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_093541_010 |
0.2362779747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_101724_010 |
0.2363150297 |
- |
- |
glutamate receptor family protein [Populus trichocarpa] |
| 20 |
Hb_006198_160 |
0.236567807 |
- |
- |
hypothetical protein POPTR_0002s24290g [Populus trichocarpa] |