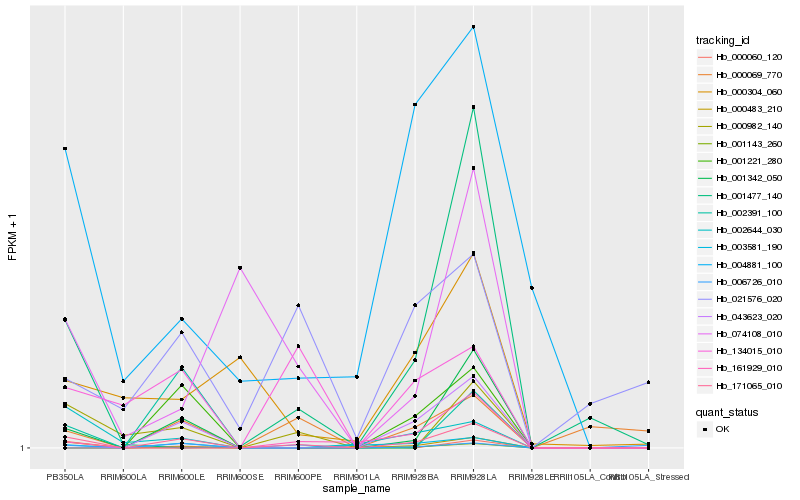
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_171065_010 |
0.0 |
- |
- |
Ribonuclease H protein [Theobroma cacao] |
| 2 |
Hb_001342_050 |
0.2527152602 |
- |
- |
- |
| 3 |
Hb_000982_140 |
0.3106509814 |
- |
- |
histidine kinase 1 plant, putative [Ricinus communis] |
| 4 |
Hb_003581_190 |
0.315754425 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_21694 [Jatropha curcas] |
| 5 |
Hb_004881_100 |
0.3464562946 |
- |
- |
Phosphoribosyltransferase family protein isoform 4 [Theobroma cacao] |
| 6 |
Hb_134015_010 |
0.3467363984 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 7 |
Hb_006726_010 |
0.3481263447 |
- |
- |
PREDICTED: uncharacterized protein K02A2.6-like [Nicotiana sylvestris] |
| 8 |
Hb_001477_140 |
0.3541743751 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000483_210 |
0.3550449402 |
- |
- |
hypothetical protein JCGZ_03497 [Jatropha curcas] |
| 10 |
Hb_002644_030 |
0.3639394571 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like, partial [Eucalyptus grandis] |
| 11 |
Hb_000069_770 |
0.3826299398 |
- |
- |
translation initiation factor [Rheum australe] |
| 12 |
Hb_043623_020 |
0.3873931646 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 13 |
Hb_001143_260 |
0.3874458542 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 14 |
Hb_021576_020 |
0.394988891 |
- |
- |
PREDICTED: uncharacterized protein LOC104426132 [Eucalyptus grandis] |
| 15 |
Hb_161929_010 |
0.3954372751 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 16 |
Hb_000304_060 |
0.4005945174 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |
| 17 |
Hb_000060_120 |
0.4008256505 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 18 |
Hb_001221_280 |
0.4024027115 |
- |
- |
- |
| 19 |
Hb_002391_100 |
0.4037322537 |
- |
- |
hypothetical protein POPTR_0007s09360g [Populus trichocarpa] |
| 20 |
Hb_074108_010 |
0.4067911413 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |