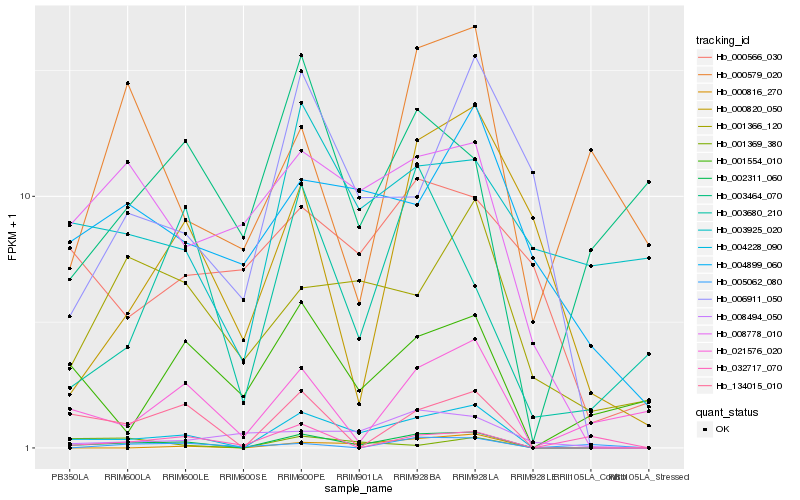
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_060 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 4 [Jatropha curcas] |
| 2 |
Hb_004228_090 |
0.1049144145 |
- |
- |
- |
| 3 |
Hb_134015_010 |
0.2365511334 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 4 |
Hb_008778_010 |
0.2988347715 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_021576_020 |
0.2997934635 |
- |
- |
PREDICTED: uncharacterized protein LOC104426132 [Eucalyptus grandis] |
| 6 |
Hb_003680_210 |
0.3076124624 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 7 |
Hb_000816_270 |
0.3099504915 |
- |
- |
- |
| 8 |
Hb_006911_050 |
0.3237748331 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 9 |
Hb_001554_010 |
0.3244297237 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 10 |
Hb_000820_050 |
0.3247276655 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 11 |
Hb_001366_120 |
0.3291424136 |
- |
- |
unknown [Zea mays] |
| 12 |
Hb_008494_050 |
0.3369352458 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 13 |
Hb_004899_060 |
0.3397051181 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 14 |
Hb_001369_380 |
0.3441685705 |
- |
- |
F-box family protein, putative [Theobroma cacao] |
| 15 |
Hb_032717_070 |
0.3471437542 |
- |
- |
PREDICTED: mucin-2-like [Jatropha curcas] |
| 16 |
Hb_005062_080 |
0.3474348322 |
- |
- |
MLO-like protein 4 [Gossypium arboreum] |
| 17 |
Hb_003464_070 |
0.3480951412 |
transcription factor |
TF Family: Trihelix |
PREDICTED: stress response protein NST1 [Jatropha curcas] |
| 18 |
Hb_000579_020 |
0.3488714375 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |
| 19 |
Hb_003925_020 |
0.3496805007 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 3 [UDP-forming]-like [Jatropha curcas] |
| 20 |
Hb_000566_030 |
0.3498459649 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |