| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
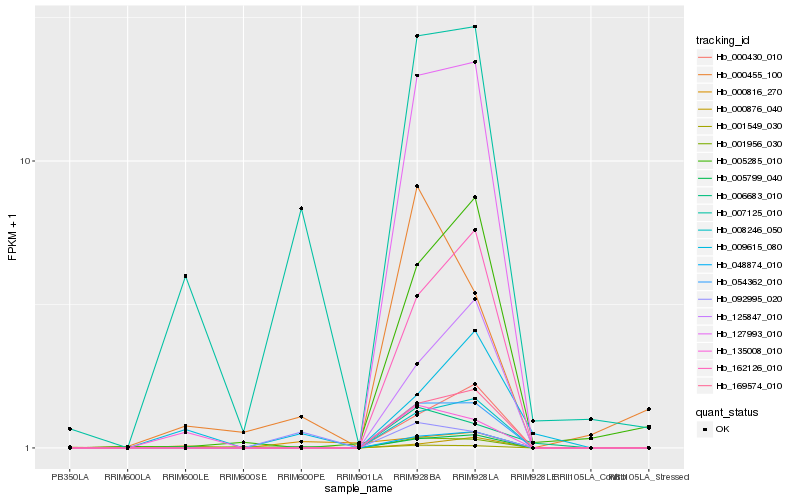
Hb_007125_010 |
0.0 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 2 |
Hb_009615_080 |
0.2491574022 |
- |
- |
hypothetical protein JCGZ_24461 [Jatropha curcas] |
| 3 |
Hb_006683_010 |
0.2795348215 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 4 |
Hb_092995_020 |
0.2822304388 |
- |
- |
- |
| 5 |
Hb_127993_010 |
0.2837961824 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 6 |
Hb_054362_010 |
0.2840504746 |
- |
- |
PREDICTED: uncharacterized protein LOC105650428 [Jatropha curcas] |
| 7 |
Hb_169574_010 |
0.2885508843 |
- |
- |
PREDICTED: uncharacterized protein LOC104120373 [Nicotiana tomentosiformis] |
| 8 |
Hb_048874_010 |
0.289006142 |
- |
- |
hypothetical protein POPTR_0016s10870g [Populus trichocarpa] |
| 9 |
Hb_005799_040 |
0.2902131578 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4-like isoform X1 [Gossypium raimondii] |
| 10 |
Hb_008246_050 |
0.2902224789 |
- |
- |
- |
| 11 |
Hb_000455_100 |
0.2904213937 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_000816_270 |
0.2905901642 |
- |
- |
- |
| 13 |
Hb_001549_030 |
0.2989851109 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_005285_010 |
0.2993990735 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000876_040 |
0.2996198889 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 16 |
Hb_135008_010 |
0.3021515415 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 17 |
Hb_162126_010 |
0.3086603034 |
- |
- |
- |
| 18 |
Hb_000430_010 |
0.3163736401 |
- |
- |
- |
| 19 |
Hb_125847_010 |
0.3234765062 |
- |
- |
- |
| 20 |
Hb_001956_030 |
0.3235778948 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Populus euphratica] |