| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
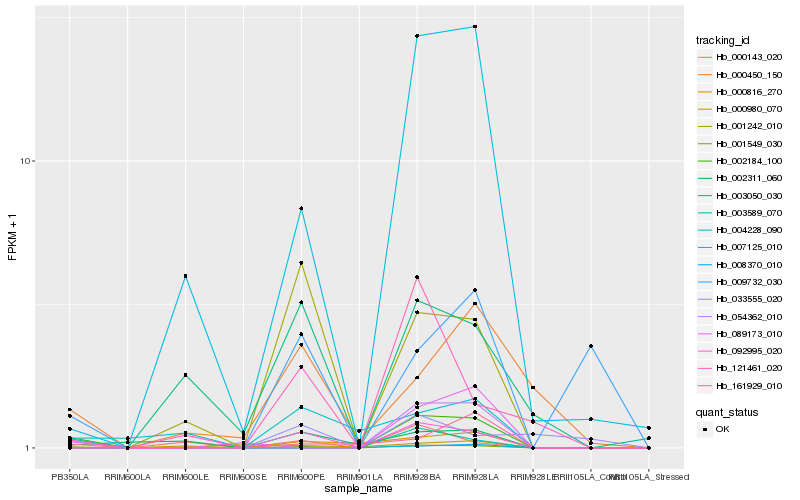
Hb_001549_030 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_092995_020 |
0.232696768 |
- |
- |
- |
| 3 |
Hb_007125_010 |
0.2989851109 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 4 |
Hb_002184_100 |
0.3232388925 |
- |
- |
hypothetical protein PRUPE_ppa020120mg [Prunus persica] |
| 5 |
Hb_001242_010 |
0.3414348538 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 6 |
Hb_121461_020 |
0.3574546467 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_009732_030 |
0.3665672701 |
- |
- |
TPA: hypothetical protein ZEAMMB73_114254 [Zea mays] |
| 8 |
Hb_003589_070 |
0.3699953958 |
- |
- |
PREDICTED: uncharacterized protein LOC104242373 [Nicotiana sylvestris] |
| 9 |
Hb_002311_060 |
0.3702035195 |
- |
- |
PREDICTED: protein trichome birefringence-like 4 [Jatropha curcas] |
| 10 |
Hb_000450_150 |
0.3709412173 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 11 |
Hb_004228_090 |
0.3742483734 |
- |
- |
- |
| 12 |
Hb_161929_010 |
0.3745758174 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 13 |
Hb_000816_270 |
0.3817184876 |
- |
- |
- |
| 14 |
Hb_033555_020 |
0.3830036068 |
- |
- |
Uncharacterized protein TCM_003129 [Theobroma cacao] |
| 15 |
Hb_089173_010 |
0.3861526485 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 16 |
Hb_003050_030 |
0.3904294997 |
- |
- |
Calcyclin-binding protein, putative [Ricinus communis] |
| 17 |
Hb_000980_070 |
0.3929567936 |
- |
- |
PREDICTED: uncharacterized protein LOC103440883 [Malus domestica] |
| 18 |
Hb_000143_020 |
0.3988142368 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 19 |
Hb_054362_010 |
0.4023930635 |
- |
- |
PREDICTED: uncharacterized protein LOC105650428 [Jatropha curcas] |
| 20 |
Hb_008370_010 |
0.4027197972 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |