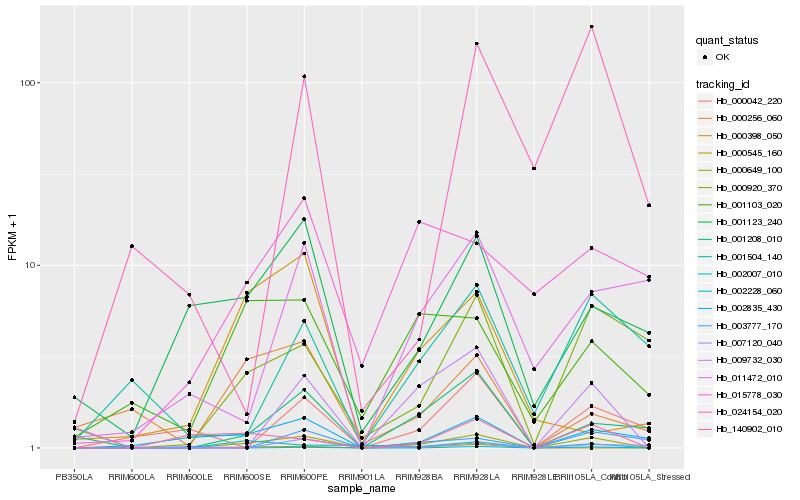
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_100 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_009732_030 |
0.3064836256 |
- |
- |
TPA: hypothetical protein ZEAMMB73_114254 [Zea mays] |
| 3 |
Hb_001208_010 |
0.3098191703 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF010-like [Jatropha curcas] |
| 4 |
Hb_002835_430 |
0.3118493004 |
- |
- |
Alliin lyase precursor, putative [Ricinus communis] |
| 5 |
Hb_000920_370 |
0.3308164631 |
- |
- |
hypothetical protein JCGZ_24527 [Jatropha curcas] |
| 6 |
Hb_140902_010 |
0.330927684 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 7 |
Hb_002228_060 |
0.3328963119 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 8 |
Hb_001103_020 |
0.3484700763 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 9 |
Hb_002007_010 |
0.352598212 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 37a [Jatropha curcas] |
| 10 |
Hb_001123_240 |
0.3536793692 |
- |
- |
JAZ10 [Hevea brasiliensis] |
| 11 |
Hb_000256_060 |
0.3584522117 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 12 |
Hb_007120_040 |
0.3763883089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000545_160 |
0.3774496844 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 14 |
Hb_000042_220 |
0.3809828577 |
- |
- |
PREDICTED: uncharacterized protein LOC105632809 [Jatropha curcas] |
| 15 |
Hb_001504_140 |
0.3823542015 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Populus euphratica] |
| 16 |
Hb_003777_170 |
0.3847482947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_015778_030 |
0.3881442698 |
- |
- |
PREDICTED: probable myosin light chain kinase DDB_G0279831 [Jatropha curcas] |
| 18 |
Hb_000398_050 |
0.3926241296 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.3 [Jatropha curcas] |
| 19 |
Hb_011472_010 |
0.3949651136 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 20 |
Hb_024154_020 |
0.3955407623 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |