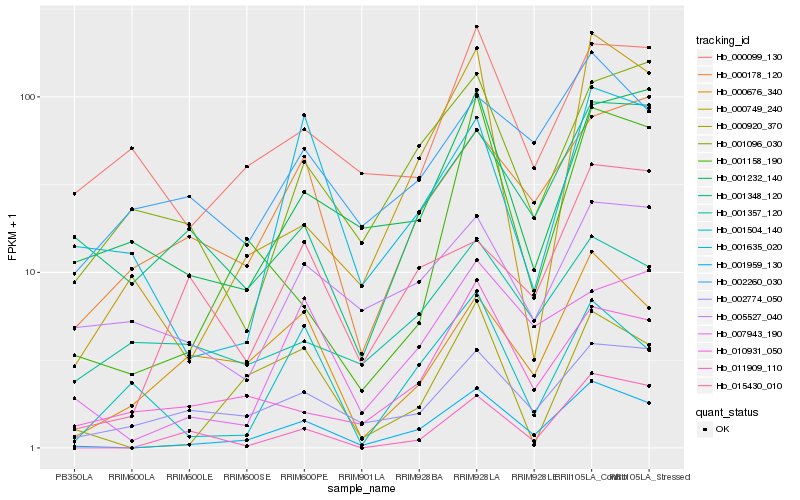
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001959_130 |
0.0 |
- |
- |
transporter, putative [Ricinus communis] |
| 2 |
Hb_000749_240 |
0.197817294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007943_190 |
0.1988331153 |
- |
- |
cytoplasmic dynein light chain, putative [Ricinus communis] |
| 4 |
Hb_000920_370 |
0.1998450586 |
- |
- |
hypothetical protein JCGZ_24527 [Jatropha curcas] |
| 5 |
Hb_002774_050 |
0.2020059138 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 6 |
Hb_011909_110 |
0.2101063239 |
- |
- |
PREDICTED: uncharacterized protein LOC105633210 [Jatropha curcas] |
| 7 |
Hb_000676_340 |
0.2101206936 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_010931_050 |
0.212599894 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 9 |
Hb_000178_120 |
0.2129732683 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 10 |
Hb_001158_190 |
0.2183249353 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET10-like [Jatropha curcas] |
| 11 |
Hb_001635_020 |
0.2229924778 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 12 |
Hb_001096_030 |
0.2234956052 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 13 |
Hb_001348_120 |
0.223756881 |
- |
- |
Phloem protein 2-B10, putative [Theobroma cacao] |
| 14 |
Hb_001232_140 |
0.2239045444 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 15 |
Hb_002260_030 |
0.2254897043 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 16 |
Hb_015430_010 |
0.2291464276 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 17 |
Hb_001504_140 |
0.2292522608 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Populus euphratica] |
| 18 |
Hb_000099_130 |
0.2352189285 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 19 |
Hb_005527_040 |
0.2376554175 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 20 |
Hb_001357_120 |
0.2384332185 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |