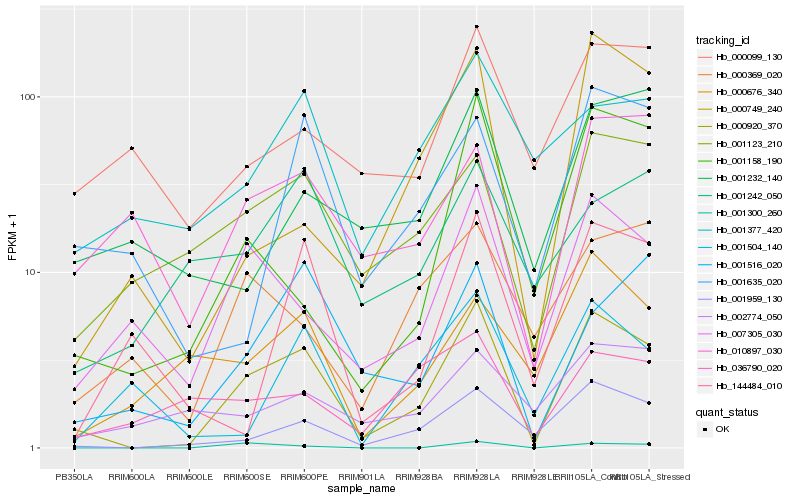
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_370 |
0.0 |
- |
- |
hypothetical protein JCGZ_24527 [Jatropha curcas] |
| 2 |
Hb_001959_130 |
0.1998450586 |
- |
- |
transporter, putative [Ricinus communis] |
| 3 |
Hb_007305_030 |
0.2287282918 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 4 |
Hb_001123_210 |
0.2433669151 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 5 |
Hb_001158_190 |
0.2534916687 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET10-like [Jatropha curcas] |
| 6 |
Hb_001300_260 |
0.2556067601 |
- |
- |
- |
| 7 |
Hb_001516_020 |
0.258364803 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-9-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_001635_020 |
0.2609944734 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 9 |
Hb_000749_240 |
0.2621305936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010897_030 |
0.2647189954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000676_340 |
0.2700772309 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000369_020 |
0.2712061838 |
- |
- |
PREDICTED: probable fructokinase-7 [Jatropha curcas] |
| 13 |
Hb_144484_010 |
0.2724106599 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 14 |
Hb_001504_140 |
0.2761300168 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Populus euphratica] |
| 15 |
Hb_000099_130 |
0.2763553945 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 16 |
Hb_001377_420 |
0.2769082451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002774_050 |
0.2813898766 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 18 |
Hb_001242_050 |
0.2815959021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_036790_020 |
0.2849862694 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |
| 20 |
Hb_001232_140 |
0.2857977324 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |