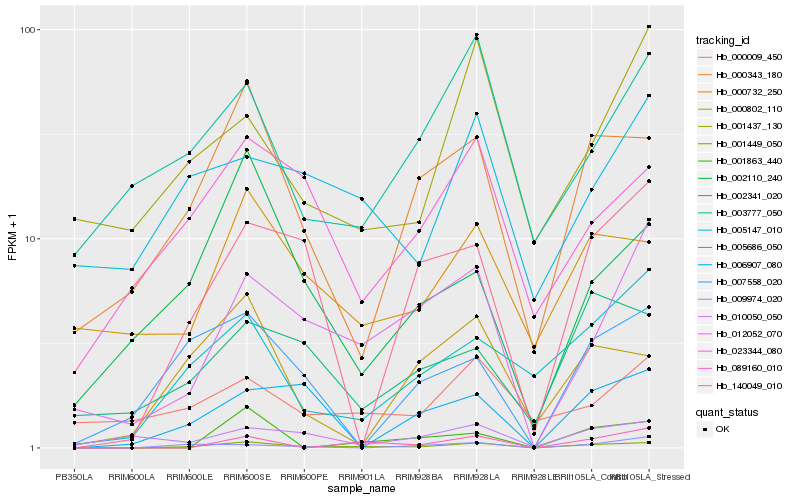
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001449_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000343_180 |
0.2278917947 |
- |
- |
NDWp3 [Podospora anserina] |
| 3 |
Hb_012052_070 |
0.2313725272 |
- |
- |
PREDICTED: uncharacterized protein LOC105635597 [Jatropha curcas] |
| 4 |
Hb_089160_010 |
0.2552964672 |
- |
- |
PREDICTED: uncharacterized protein LOC105793196 [Gossypium raimondii] |
| 5 |
Hb_005147_010 |
0.2574290473 |
- |
- |
PREDICTED: uncharacterized protein LOC105629670 [Jatropha curcas] |
| 6 |
Hb_140049_010 |
0.2580927485 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 7 |
Hb_023344_080 |
0.2585815414 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000732_250 |
0.2626112182 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B2, mitochondrial-like [Populus euphratica] |
| 9 |
Hb_000802_110 |
0.2633999388 |
- |
- |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 10 |
Hb_002110_240 |
0.2659510563 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 11 |
Hb_001863_440 |
0.2661031578 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 12 |
Hb_007558_020 |
0.2714908205 |
- |
- |
hypothetical protein JCGZ_17631 [Jatropha curcas] |
| 13 |
Hb_006907_080 |
0.2735692703 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 14 |
Hb_003777_050 |
0.2740823368 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 15 |
Hb_002341_020 |
0.2760602375 |
- |
- |
PREDICTED: pyruvate decarboxylase 2 [Populus euphratica] |
| 16 |
Hb_005686_050 |
0.2788156579 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 17 |
Hb_009974_020 |
0.2823405706 |
- |
- |
PREDICTED: uncharacterized protein LOC104427753 [Eucalyptus grandis] |
| 18 |
Hb_010050_050 |
0.2845535595 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_000009_450 |
0.2855286301 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_001437_130 |
0.2855821793 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |