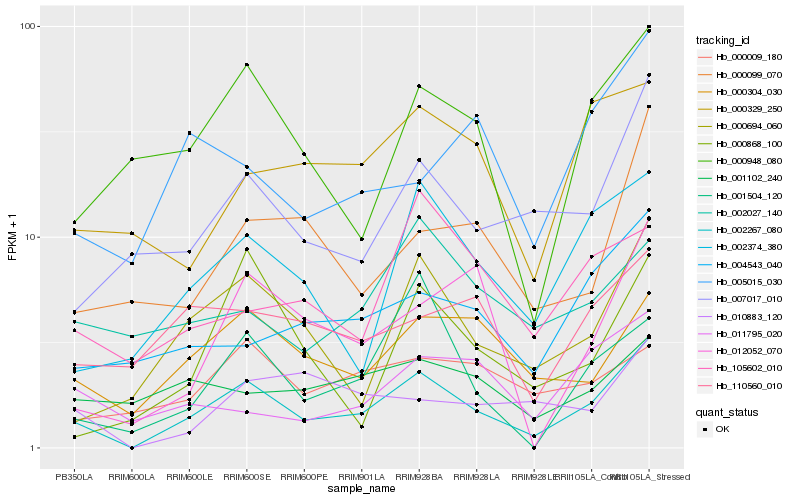
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002267_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635094 [Jatropha curcas] |
| 2 |
Hb_000694_060 |
0.1931127421 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 3 |
Hb_002374_380 |
0.2037244546 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_007017_010 |
0.2094197118 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 5 |
Hb_000304_030 |
0.209657615 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 6 |
Hb_000948_080 |
0.2160249923 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 7 |
Hb_004543_040 |
0.2262965734 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_001504_120 |
0.2305505223 |
- |
- |
- |
| 9 |
Hb_012052_070 |
0.2320504754 |
- |
- |
PREDICTED: uncharacterized protein LOC105635597 [Jatropha curcas] |
| 10 |
Hb_000099_070 |
0.2332534175 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 11 |
Hb_110560_010 |
0.2354717012 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000329_250 |
0.237002355 |
- |
- |
PREDICTED: vesicle-associated protein 1-3 isoform X1 [Fragaria vesca subsp. vesca] |
| 13 |
Hb_010883_120 |
0.2399097766 |
- |
- |
- |
| 14 |
Hb_000868_100 |
0.2411088909 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF144A [Jatropha curcas] |
| 15 |
Hb_000009_180 |
0.2434472398 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 16 |
Hb_005015_030 |
0.245952453 |
- |
- |
PREDICTED: uncharacterized protein LOC105636546 [Jatropha curcas] |
| 17 |
Hb_011795_020 |
0.2465306984 |
- |
- |
- |
| 18 |
Hb_105602_010 |
0.2473592693 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 19 |
Hb_001102_240 |
0.247620914 |
- |
- |
PREDICTED: NADH kinase isoform X1 [Jatropha curcas] |
| 20 |
Hb_002027_140 |
0.2480805065 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |