| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
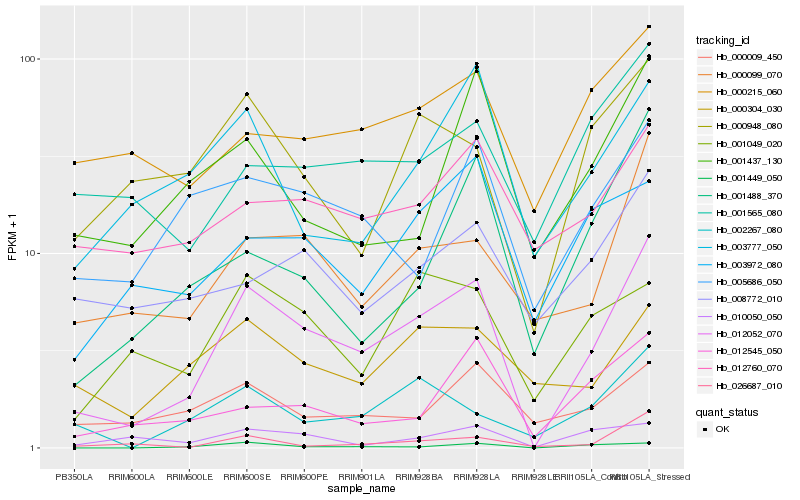
Hb_012052_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635597 [Jatropha curcas] |
| 2 |
Hb_000099_070 |
0.2109214465 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 3 |
Hb_001449_050 |
0.2313725272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002267_080 |
0.2320504754 |
- |
- |
PREDICTED: uncharacterized protein LOC105635094 [Jatropha curcas] |
| 5 |
Hb_000948_080 |
0.2356119324 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 6 |
Hb_005686_050 |
0.2384944292 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 7 |
Hb_001488_370 |
0.2401673948 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 8 |
Hb_000304_030 |
0.2419586938 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 9 |
Hb_012545_050 |
0.2431269903 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Jatropha curcas] |
| 10 |
Hb_001049_020 |
0.2441448363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_026687_010 |
0.2447398583 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Eucalyptus grandis] |
| 12 |
Hb_003777_050 |
0.244951111 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 13 |
Hb_012760_070 |
0.2455545415 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_001565_080 |
0.2462329623 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM17-2-like [Jatropha curcas] |
| 15 |
Hb_000009_450 |
0.2464891687 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_010050_050 |
0.2465730365 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 17 |
Hb_000215_060 |
0.2475981532 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM17-2-like [Jatropha curcas] |
| 18 |
Hb_003972_080 |
0.2484411759 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 19 |
Hb_001437_130 |
0.2489755874 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |
| 20 |
Hb_008772_010 |
0.2499732335 |
- |
- |
conserved hypothetical protein [Ricinus communis] |