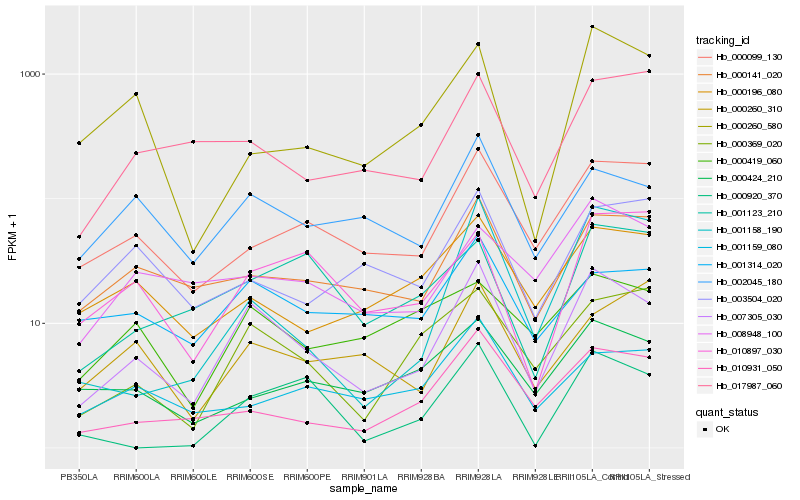
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007305_030 |
0.0 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 2 |
Hb_002045_180 |
0.1877154308 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 3 |
Hb_000369_020 |
0.1915922953 |
- |
- |
PREDICTED: probable fructokinase-7 [Jatropha curcas] |
| 4 |
Hb_000141_020 |
0.1919900444 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 6 [Pyrus x bretschneideri] |
| 5 |
Hb_000099_130 |
0.192136603 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 6 |
Hb_010931_050 |
0.1956315386 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 7 |
Hb_001314_020 |
0.2075351667 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_017987_060 |
0.2124490542 |
- |
- |
hypothetical protein JCGZ_15971 [Jatropha curcas] |
| 9 |
Hb_000196_080 |
0.2143007785 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_008948_100 |
0.2184329683 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 11 |
Hb_001158_190 |
0.2199779437 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET10-like [Jatropha curcas] |
| 12 |
Hb_000424_210 |
0.2205528759 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000419_060 |
0.2239754091 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |
| 14 |
Hb_010897_030 |
0.2251518607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001123_210 |
0.2258152623 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 16 |
Hb_001159_080 |
0.2263918822 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 17 |
Hb_000260_580 |
0.2269717501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000920_370 |
0.2287282918 |
- |
- |
hypothetical protein JCGZ_24527 [Jatropha curcas] |
| 19 |
Hb_000260_310 |
0.2289486481 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 20 |
Hb_003504_020 |
0.2289790146 |
- |
- |
PREDICTED: uncharacterized protein LOC105647732 [Jatropha curcas] |