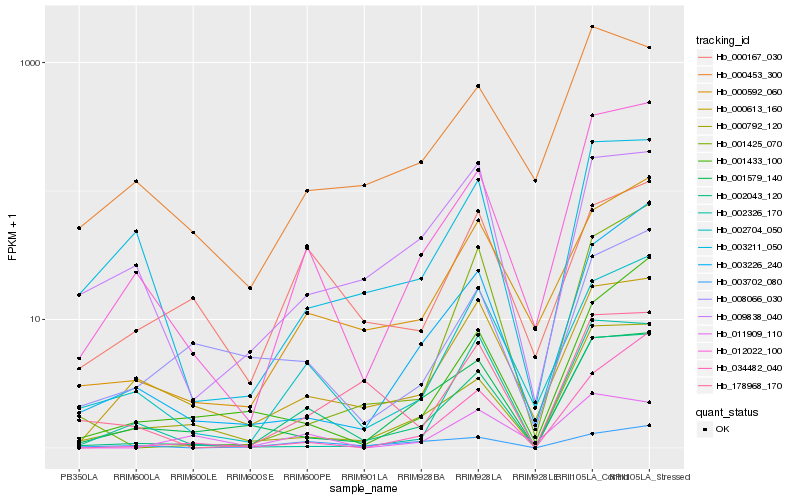
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002043_120 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_012022_100 |
0.1215794691 |
- |
- |
CPRD49 family protein [Populus trichocarpa] |
| 3 |
Hb_002704_050 |
0.1581197806 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |
| 4 |
Hb_000792_120 |
0.1707194566 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000592_060 |
0.1720425691 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 6 |
Hb_000613_160 |
0.1881906377 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 7 |
Hb_001579_140 |
0.1992169031 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000453_300 |
0.1992307058 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 9 |
Hb_003226_240 |
0.202706829 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 10 |
Hb_178968_170 |
0.2050378993 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 homolog [Jatropha curcas] |
| 11 |
Hb_001425_070 |
0.2050648856 |
- |
- |
- |
| 12 |
Hb_003211_050 |
0.2056104307 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 13 |
Hb_008066_030 |
0.211516796 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 14 |
Hb_000167_030 |
0.2116366837 |
- |
- |
PREDICTED: uncharacterized protein LOC105641111 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011909_110 |
0.2123197359 |
- |
- |
PREDICTED: uncharacterized protein LOC105633210 [Jatropha curcas] |
| 16 |
Hb_034482_040 |
0.213910971 |
- |
- |
aspartate kinase, partial [Francisella tularensis] |
| 17 |
Hb_001433_100 |
0.2174265637 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002326_170 |
0.2174450531 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 19 |
Hb_003702_080 |
0.2191038614 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 20 |
Hb_009838_040 |
0.2194397782 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |