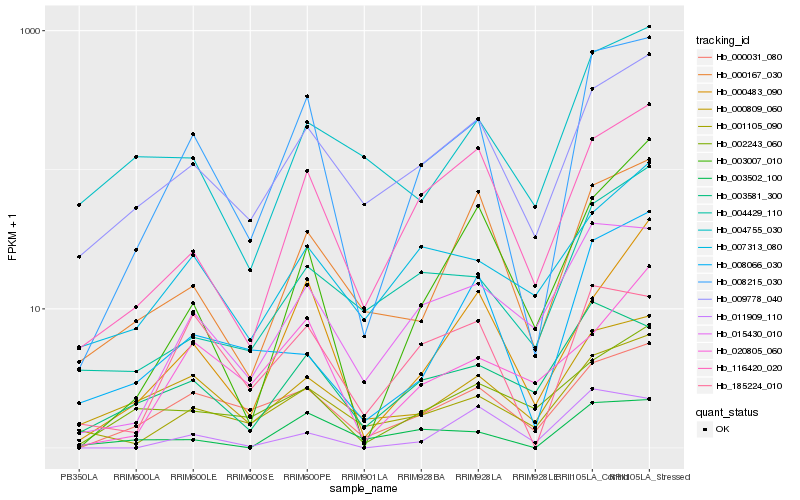
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008215_030 |
0.0 |
- |
- |
hypothetical protein 3 [Hevea brasiliensis] |
| 2 |
Hb_009778_040 |
0.1797786008 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 3 |
Hb_015430_010 |
0.1811781366 |
- |
- |
PREDICTED: uncharacterized protein LOC105125506 isoform X2 [Populus euphratica] |
| 4 |
Hb_001105_090 |
0.1841168353 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_116420_020 |
0.1850148818 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 6 |
Hb_000031_080 |
0.1870979028 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 7 |
Hb_000809_060 |
0.1979569254 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 8 |
Hb_000483_090 |
0.2019909999 |
- |
- |
- |
| 9 |
Hb_000167_030 |
0.2033797939 |
- |
- |
PREDICTED: uncharacterized protein LOC105641111 isoform X1 [Jatropha curcas] |
| 10 |
Hb_185224_010 |
0.207525088 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_003502_100 |
0.2104751274 |
- |
- |
- |
| 12 |
Hb_008066_030 |
0.2120234182 |
- |
- |
PREDICTED: uncharacterized protein LOC105631333 [Jatropha curcas] |
| 13 |
Hb_004429_110 |
0.2152545494 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 14 |
Hb_003007_010 |
0.2167851194 |
- |
- |
PREDICTED: probable peptide/nitrate transporter At3g43790 isoform X1 [Populus euphratica] |
| 15 |
Hb_003581_300 |
0.2216289831 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002243_060 |
0.2239524077 |
- |
- |
hypothetical protein POPTR_0022s00750g [Populus trichocarpa] |
| 17 |
Hb_004755_030 |
0.2266008395 |
- |
- |
EG964 [Manihot glaziovii] |
| 18 |
Hb_011909_110 |
0.2284683844 |
- |
- |
PREDICTED: uncharacterized protein LOC105633210 [Jatropha curcas] |
| 19 |
Hb_020805_060 |
0.2293093595 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 20 |
Hb_007313_080 |
0.2294102323 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |