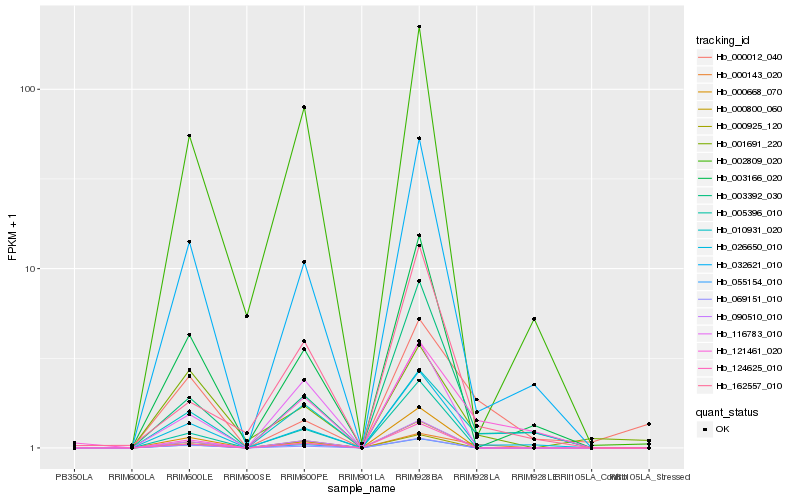
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000143_020 |
0.0 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 2 |
Hb_010931_020 |
0.2135541676 |
- |
- |
Kunitz-type protease inhibitor KPI-F4 [Populus trichocarpa x Populus deltoides] |
| 3 |
Hb_026650_010 |
0.2259732555 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 4 |
Hb_032621_010 |
0.2432758281 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 5 |
Hb_003392_030 |
0.2465379054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000012_040 |
0.2509934283 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 7 |
Hb_162557_010 |
0.2549638228 |
- |
- |
PREDICTED: uncharacterized protein LOC105634192 [Jatropha curcas] |
| 8 |
Hb_090510_010 |
0.2568809383 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 9 |
Hb_055154_010 |
0.2571440252 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 10 |
Hb_121461_020 |
0.2586699489 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000800_060 |
0.2595278414 |
- |
- |
hypothetical protein POPTR_0008s02870g [Populus trichocarpa] |
| 12 |
Hb_005396_010 |
0.2651396799 |
- |
- |
PREDICTED: interleukin enhancer-binding factor 3-B-like [Jatropha curcas] |
| 13 |
Hb_069151_010 |
0.2701351542 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Gossypium raimondii] |
| 14 |
Hb_000925_120 |
0.2704239794 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 15 |
Hb_116783_010 |
0.2708655798 |
- |
- |
hypothetical protein POPTR_0011s16190g [Populus trichocarpa] |
| 16 |
Hb_002809_020 |
0.2723902219 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 17 |
Hb_124625_010 |
0.2739347637 |
- |
- |
hypothetical protein JCGZ_01142 [Jatropha curcas] |
| 18 |
Hb_001691_220 |
0.2785085468 |
- |
- |
PREDICTED: uncharacterized protein LOC105649537 [Jatropha curcas] |
| 19 |
Hb_003166_020 |
0.2799971584 |
- |
- |
PREDICTED: RNA-binding E3 ubiquitin-protein ligase MEX3C-like [Jatropha curcas] |
| 20 |
Hb_000668_070 |
0.2800011368 |
- |
- |
conserved hypothetical protein [Ricinus communis] |