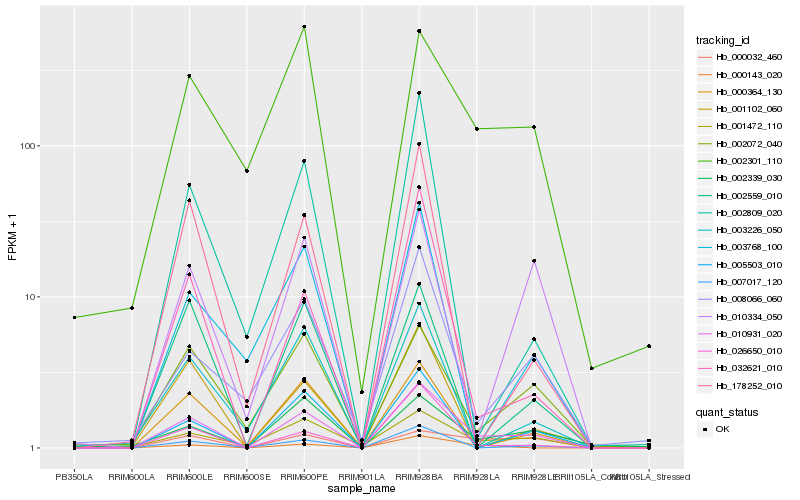
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010931_020 |
0.0 |
- |
- |
Kunitz-type protease inhibitor KPI-F4 [Populus trichocarpa x Populus deltoides] |
| 2 |
Hb_005503_010 |
0.1941723772 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 3 |
Hb_007017_120 |
0.1945648211 |
- |
- |
PREDICTED: exocyst complex component EXO84A [Jatropha curcas] |
| 4 |
Hb_000364_130 |
0.1969443404 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_001472_110 |
0.2004082799 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 6 |
Hb_026650_010 |
0.2053638823 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 7 |
Hb_002339_030 |
0.2061348076 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_008066_060 |
0.2073901702 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 9 |
Hb_002072_040 |
0.2095876541 |
- |
- |
PREDICTED: uncharacterized protein LOC105630881 [Jatropha curcas] |
| 10 |
Hb_032621_010 |
0.2096988188 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 11 |
Hb_001102_060 |
0.2129916618 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At1g78530 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000143_020 |
0.2135541676 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 13 |
Hb_000032_460 |
0.2155396175 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 14 |
Hb_178252_010 |
0.2176191572 |
- |
- |
hypothetical protein POPTR_0018s01890g [Populus trichocarpa] |
| 15 |
Hb_002301_110 |
0.2221531797 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 16 |
Hb_003226_050 |
0.2245738115 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 17 |
Hb_002559_010 |
0.2253480144 |
- |
- |
hypothetical protein JCGZ_06274 [Jatropha curcas] |
| 18 |
Hb_002809_020 |
0.2271892285 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 19 |
Hb_003768_100 |
0.2295014813 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 20 |
Hb_010334_050 |
0.2313016287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |