| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
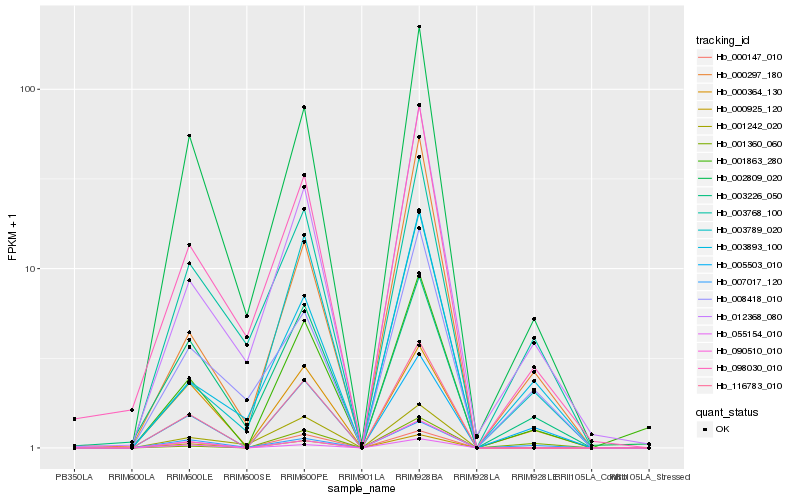
Hb_001863_280 |
0.0 |
- |
- |
PREDICTED: protein EXORDIUM-like 2 [Jatropha curcas] |
| 2 |
Hb_116783_010 |
0.1403391262 |
- |
- |
hypothetical protein POPTR_0011s16190g [Populus trichocarpa] |
| 3 |
Hb_005503_010 |
0.1514227843 |
- |
- |
PREDICTED: probable protein Pop3 [Jatropha curcas] |
| 4 |
Hb_002809_020 |
0.1546620181 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 5 |
Hb_000925_120 |
0.1568672598 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 6 |
Hb_012368_080 |
0.1639065585 |
- |
- |
PREDICTED: uncharacterized protein LOC105126277 [Populus euphratica] |
| 7 |
Hb_098030_010 |
0.1680974711 |
- |
- |
hypothetical protein JCGZ_06251 [Jatropha curcas] |
| 8 |
Hb_000147_010 |
0.1763359123 |
- |
- |
PREDICTED: UDP-glycosyltransferase 87A1-like [Jatropha curcas] |
| 9 |
Hb_007017_120 |
0.1795023253 |
- |
- |
PREDICTED: exocyst complex component EXO84A [Jatropha curcas] |
| 10 |
Hb_003789_020 |
0.1802533401 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 11 |
Hb_003768_100 |
0.182806638 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 12 |
Hb_055154_010 |
0.1830430582 |
- |
- |
PREDICTED: laccase-7 [Jatropha curcas] |
| 13 |
Hb_003226_050 |
0.1866854852 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 14 |
Hb_000297_180 |
0.1881817894 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 15 |
Hb_000364_130 |
0.1911080935 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_003893_100 |
0.1936624382 |
- |
- |
PREDICTED: uncharacterized protein LOC105648774 [Jatropha curcas] |
| 17 |
Hb_090510_010 |
0.1938124588 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 18 |
Hb_001242_020 |
0.1950353627 |
- |
- |
hypothetical protein JCGZ_08334 [Jatropha curcas] |
| 19 |
Hb_001360_060 |
0.1953539854 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 20 |
Hb_008418_010 |
0.1976265244 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |