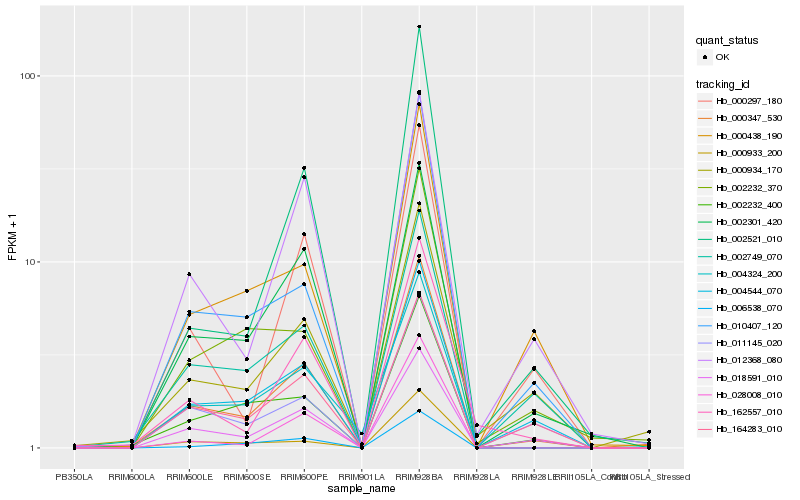
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_530 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 2 |
Hb_018591_010 |
0.109603885 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_004544_070 |
0.1124266619 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000934_170 |
0.1138985522 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_002301_420 |
0.1174639098 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 6 |
Hb_002749_070 |
0.1178270094 |
- |
- |
PREDICTED: uncharacterized protein LOC105635941 [Jatropha curcas] |
| 7 |
Hb_010407_120 |
0.1215244396 |
- |
- |
PREDICTED: glutathione transferase GST 23-like [Jatropha curcas] |
| 8 |
Hb_002232_400 |
0.1216872123 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 9 |
Hb_002521_010 |
0.1259071211 |
- |
- |
cinnamate 4-hydroxylase, putative [Ricinus communis] |
| 10 |
Hb_000438_190 |
0.1279156701 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 11 |
Hb_164283_010 |
0.1288679731 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 12 |
Hb_028008_010 |
0.1294727752 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23 [Jatropha curcas] |
| 13 |
Hb_000297_180 |
0.1313729554 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 14 |
Hb_006538_070 |
0.1330222085 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 15 |
Hb_011145_020 |
0.1332507262 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 16 |
Hb_162557_010 |
0.1338833213 |
- |
- |
PREDICTED: uncharacterized protein LOC105634192 [Jatropha curcas] |
| 17 |
Hb_012368_080 |
0.1346299627 |
- |
- |
PREDICTED: uncharacterized protein LOC105126277 [Populus euphratica] |
| 18 |
Hb_002232_370 |
0.1349984839 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 19 |
Hb_000933_200 |
0.1363969206 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 20 |
Hb_004324_200 |
0.1379683208 |
- |
- |
PREDICTED: transcription factor TRY-like [Populus euphratica] |