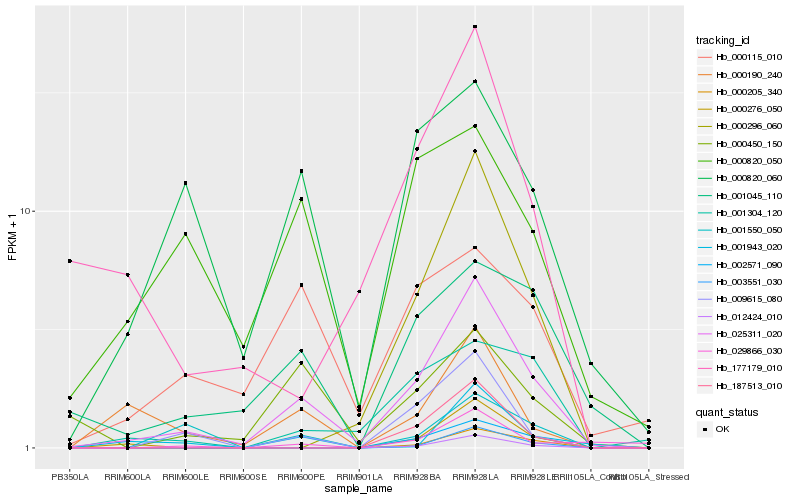
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025311_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105039527 [Elaeis guineensis] |
| 2 |
Hb_009615_080 |
0.218357206 |
- |
- |
hypothetical protein JCGZ_24461 [Jatropha curcas] |
| 3 |
Hb_012424_010 |
0.2539323426 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 4 |
Hb_000820_060 |
0.2602355591 |
- |
- |
- |
| 5 |
Hb_001045_110 |
0.2607194969 |
transcription factor |
TF Family: M-type |
flowering locus C-like protein [Juglans regia] |
| 6 |
Hb_000190_240 |
0.2635294976 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_000296_060 |
0.2654090495 |
- |
- |
- |
| 8 |
Hb_001550_050 |
0.2672902082 |
- |
- |
PREDICTED: J domain-containing protein spf31 [Eucalyptus grandis] |
| 9 |
Hb_000450_150 |
0.2744792906 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 10 |
Hb_001304_120 |
0.2758285337 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 11 |
Hb_001943_020 |
0.277210978 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase-like, partial [Populus euphratica] |
| 12 |
Hb_177179_010 |
0.2777682349 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 13 |
Hb_000276_050 |
0.2793917686 |
- |
- |
- |
| 14 |
Hb_000115_010 |
0.2794319355 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_002571_090 |
0.2821988162 |
- |
- |
- |
| 16 |
Hb_000820_050 |
0.2849102846 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 17 |
Hb_187513_010 |
0.2851925784 |
- |
- |
PREDICTED: uncharacterized protein LOC105761922 [Gossypium raimondii] |
| 18 |
Hb_000205_340 |
0.2894311023 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |
| 19 |
Hb_029866_030 |
0.2918804445 |
- |
- |
hypothetical protein CICLE_v10006898mg [Citrus clementina] |
| 20 |
Hb_003551_030 |
0.294542338 |
- |
- |
hypothetical protein POPTR_0016s03850g [Populus trichocarpa] |