| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
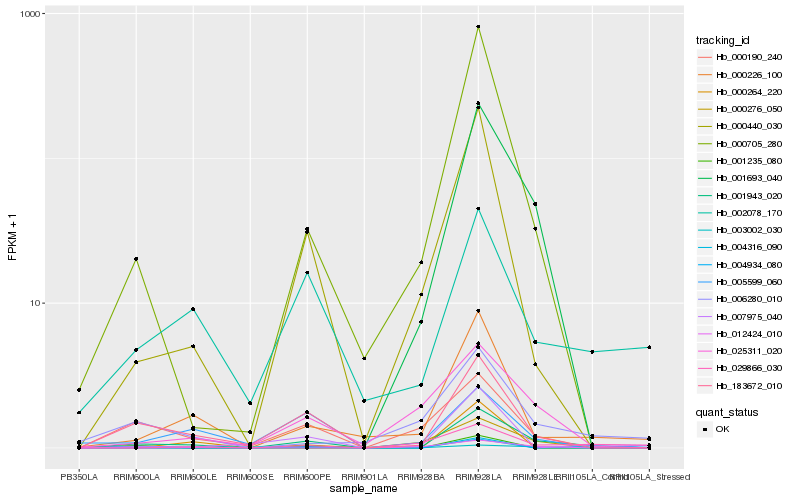
Hb_001943_020 |
0.0 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase-like, partial [Populus euphratica] |
| 2 |
Hb_183672_010 |
0.1976652832 |
- |
- |
PREDICTED: (S)-tetrahydroprotoberberine N-methyltransferase isoform X3 [Jatropha curcas] |
| 3 |
Hb_000226_100 |
0.2506600211 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 4 |
Hb_000440_030 |
0.2636139215 |
- |
- |
50 kDa protein [Hevea brasiliensis] |
| 5 |
Hb_000705_280 |
0.2646844065 |
- |
- |
PREDICTED: uncharacterized protein LOC104601116 [Nelumbo nucifera] |
| 6 |
Hb_000190_240 |
0.2659705985 |
- |
- |
unnamed protein product [Coffea canephora] |
| 7 |
Hb_002078_170 |
0.2746675199 |
- |
- |
ELMO/CED-12 domain-containing protein isoform 3 [Theobroma cacao] |
| 8 |
Hb_025311_020 |
0.277210978 |
- |
- |
PREDICTED: uncharacterized protein LOC105039527 [Elaeis guineensis] |
| 9 |
Hb_006280_010 |
0.2907703967 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_007975_040 |
0.3079126679 |
- |
- |
PREDICTED: uncharacterized protein LOC104610918 [Nelumbo nucifera] |
| 11 |
Hb_001235_080 |
0.3140416439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005599_060 |
0.3170061773 |
- |
- |
PREDICTED: syntaxin-72 isoform X1 [Jatropha curcas] |
| 13 |
Hb_029866_030 |
0.3269738317 |
- |
- |
hypothetical protein CICLE_v10006898mg [Citrus clementina] |
| 14 |
Hb_003002_030 |
0.334678464 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_000264_220 |
0.3388033135 |
- |
- |
PREDICTED: uncharacterized protein LOC105649527 [Jatropha curcas] |
| 16 |
Hb_000276_050 |
0.3408456644 |
- |
- |
- |
| 17 |
Hb_001693_040 |
0.3413048589 |
- |
- |
- |
| 18 |
Hb_004934_080 |
0.3480523468 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 19 |
Hb_012424_010 |
0.3489218638 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 20 |
Hb_004316_090 |
0.3491144253 |
- |
- |
hypothetical protein JCGZ_21197 [Jatropha curcas] |