| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
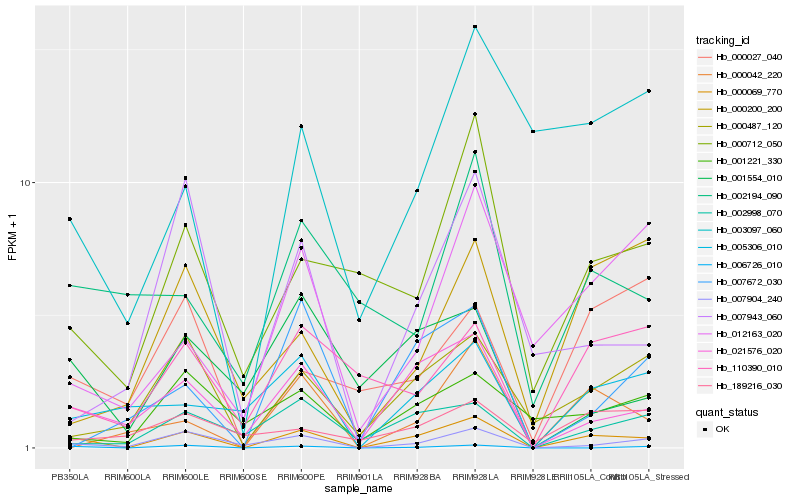
Hb_000069_770 |
0.0 |
- |
- |
translation initiation factor [Rheum australe] |
| 2 |
Hb_021576_020 |
0.2249951317 |
- |
- |
PREDICTED: uncharacterized protein LOC104426132 [Eucalyptus grandis] |
| 3 |
Hb_007904_240 |
0.2448663452 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |
| 4 |
Hb_006726_010 |
0.2817684937 |
- |
- |
PREDICTED: uncharacterized protein K02A2.6-like [Nicotiana sylvestris] |
| 5 |
Hb_000487_120 |
0.2827816027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002998_070 |
0.2911513867 |
- |
- |
hypothetical protein CARUB_v10019139mg, partial [Capsella rubella] |
| 7 |
Hb_005306_010 |
0.2941202673 |
- |
- |
- |
| 8 |
Hb_000712_050 |
0.2945238599 |
- |
- |
PREDICTED: inositol oxygenase 1-like isoform X1 [Citrus sinensis] |
| 9 |
Hb_000042_220 |
0.3019458519 |
- |
- |
PREDICTED: uncharacterized protein LOC105632809 [Jatropha curcas] |
| 10 |
Hb_110390_010 |
0.3077527057 |
- |
- |
hypothetical protein JCGZ_19741 [Jatropha curcas] |
| 11 |
Hb_012163_020 |
0.3108658448 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X3 [Jatropha curcas] |
| 12 |
Hb_001554_010 |
0.3120430622 |
- |
- |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 13 |
Hb_007943_060 |
0.3130887624 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 14 |
Hb_001221_330 |
0.3190135408 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 15 |
Hb_003097_060 |
0.3192184532 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 16 |
Hb_000027_040 |
0.3244124282 |
- |
- |
PREDICTED: uncharacterized protein LOC105636980 [Jatropha curcas] |
| 17 |
Hb_002194_090 |
0.3256876647 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_007672_030 |
0.3259250597 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 19 |
Hb_000200_200 |
0.3290369706 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 20 |
Hb_189216_030 |
0.3302784072 |
- |
- |
Pleiotropic drug resistance protein, putative [Ricinus communis] |