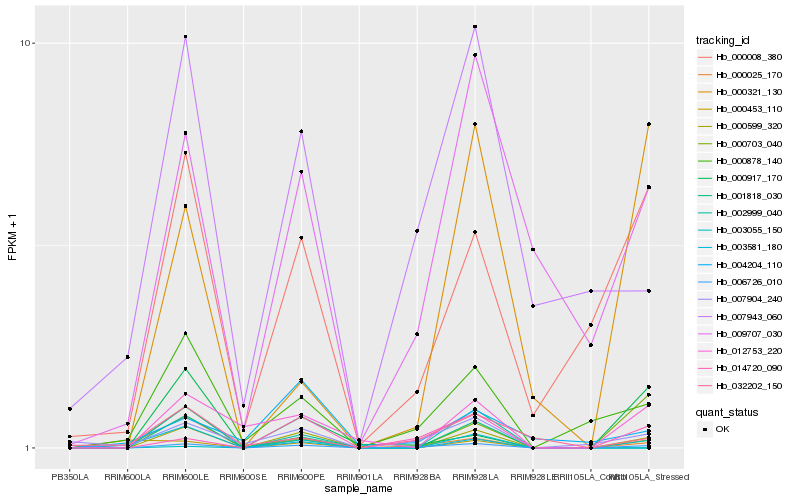
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_320 |
0.0 |
- |
- |
hypothetical protein JCGZ_17662 [Jatropha curcas] |
| 2 |
Hb_012753_220 |
0.2772446047 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 3 |
Hb_000025_170 |
0.2933074706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000008_380 |
0.2959592042 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000878_140 |
0.2969468666 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 6 |
Hb_000703_040 |
0.3116584501 |
- |
- |
hypothetical protein RCOM_1177160 [Ricinus communis] |
| 7 |
Hb_001818_030 |
0.3179125099 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 8 |
Hb_009707_030 |
0.3207419863 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620 [Jatropha curcas] |
| 9 |
Hb_014720_090 |
0.3214181931 |
- |
- |
- |
| 10 |
Hb_003055_150 |
0.3227659835 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 11 |
Hb_000917_170 |
0.3243325394 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 12 |
Hb_002999_040 |
0.3258101785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007904_240 |
0.3304812698 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |
| 14 |
Hb_006726_010 |
0.3322098714 |
- |
- |
PREDICTED: uncharacterized protein K02A2.6-like [Nicotiana sylvestris] |
| 15 |
Hb_000321_130 |
0.3581687523 |
- |
- |
Alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_007943_060 |
0.3599630679 |
- |
- |
Rop5 [Hevea brasiliensis] |
| 17 |
Hb_004204_110 |
0.3614732992 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 18 |
Hb_000453_110 |
0.3738552016 |
- |
- |
PREDICTED: alkaline/neutral invertase CINV2-like isoform X1 [Citrus sinensis] |
| 19 |
Hb_032202_150 |
0.3754087564 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR14 [Jatropha curcas] |
| 20 |
Hb_003581_180 |
0.3766045691 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |