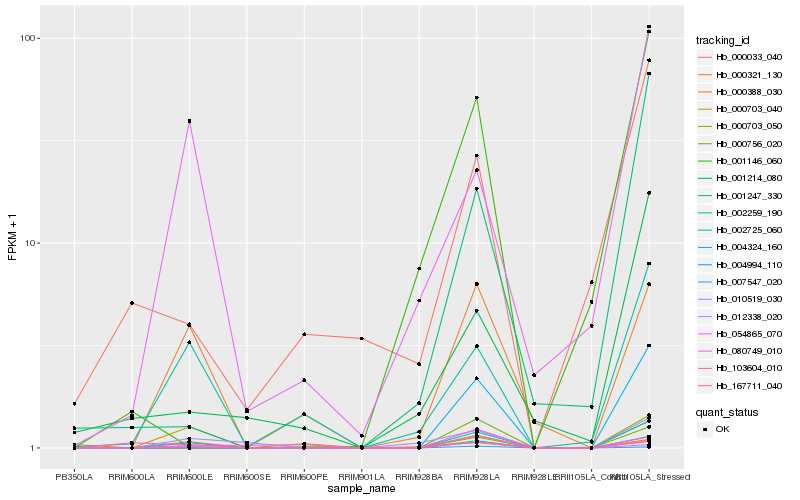
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004324_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 2 |
Hb_000321_130 |
0.2018596453 |
- |
- |
Alcohol dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_007547_020 |
0.211244673 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase A-like [Populus euphratica] |
| 4 |
Hb_002259_190 |
0.2178980946 |
- |
- |
Omega-3 fatty acid desaturase, endoplasmic reticulum, putative [Ricinus communis] |
| 5 |
Hb_000703_040 |
0.222418157 |
- |
- |
hypothetical protein RCOM_1177160 [Ricinus communis] |
| 6 |
Hb_012338_020 |
0.2277341411 |
- |
- |
- |
| 7 |
Hb_054865_070 |
0.2516178597 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 8 |
Hb_010519_030 |
0.2791450565 |
- |
- |
- |
| 9 |
Hb_000388_030 |
0.3210205513 |
- |
- |
hypothetical protein JCGZ_13427 [Jatropha curcas] |
| 10 |
Hb_002725_060 |
0.3394916169 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 11 |
Hb_004994_110 |
0.3396511539 |
- |
- |
- |
| 12 |
Hb_167711_040 |
0.3487618598 |
- |
- |
PREDICTED: cucumisin-like [Jatropha curcas] |
| 13 |
Hb_000703_050 |
0.3639566674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_103604_010 |
0.3709597738 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 15 |
Hb_001247_330 |
0.3721069537 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 16 |
Hb_080749_010 |
0.377706759 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Populus euphratica] |
| 17 |
Hb_001214_080 |
0.3779859454 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38)-like [Jatropha curcas] |
| 18 |
Hb_000033_040 |
0.3785451065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000756_020 |
0.3797724636 |
- |
- |
BnaA05g19440D [Brassica napus] |
| 20 |
Hb_001146_060 |
0.3814186588 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |