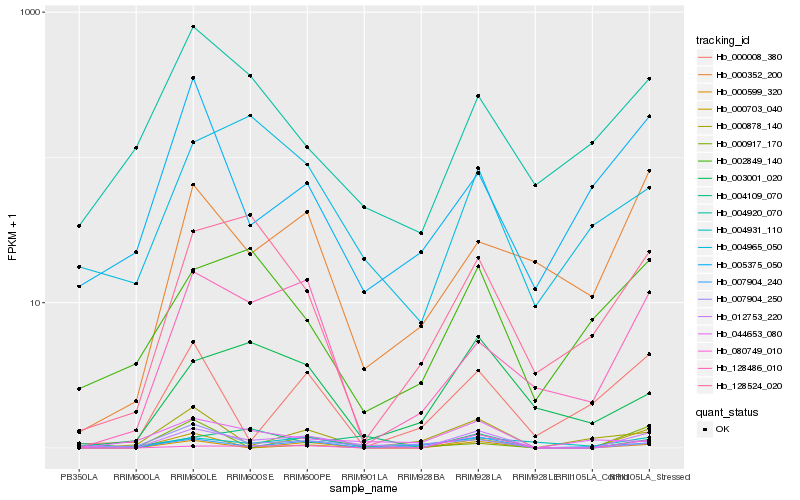
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012753_220 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 2 |
Hb_000599_320 |
0.2772446047 |
- |
- |
hypothetical protein JCGZ_17662 [Jatropha curcas] |
| 3 |
Hb_128486_010 |
0.2988899969 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 4 |
Hb_000352_200 |
0.3185068018 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 5 |
Hb_007904_250 |
0.3227757995 |
- |
- |
PREDICTED: vignain-like [Jatropha curcas] |
| 6 |
Hb_000008_380 |
0.3368866133 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_000703_040 |
0.3371371067 |
- |
- |
hypothetical protein RCOM_1177160 [Ricinus communis] |
| 8 |
Hb_128524_020 |
0.3380486531 |
- |
- |
PREDICTED: aspartic proteinase-like protein 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003001_020 |
0.3437802908 |
- |
- |
hypothetical protein JCGZ_17508 [Jatropha curcas] |
| 10 |
Hb_007904_240 |
0.3444338841 |
- |
- |
PREDICTED: protein terminal ear1 [Jatropha curcas] |
| 11 |
Hb_080749_010 |
0.3456476563 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Populus euphratica] |
| 12 |
Hb_000917_170 |
0.3491689133 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 13 |
Hb_000878_140 |
0.3493664881 |
- |
- |
PREDICTED: cell division cycle 20.2, cofactor of APC complex-like [Jatropha curcas] |
| 14 |
Hb_004931_110 |
0.3536987418 |
- |
- |
inorganic phosphate transporter, putative [Ricinus communis] |
| 15 |
Hb_002849_140 |
0.3556299113 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 16 |
Hb_004920_070 |
0.3588986354 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 17 |
Hb_004965_050 |
0.3615850215 |
- |
- |
PREDICTED: uncharacterized protein LOC105645055 [Jatropha curcas] |
| 18 |
Hb_005375_050 |
0.3640352839 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 19 |
Hb_044653_080 |
0.3649060013 |
- |
- |
Uncharacterized protein TCM_045144 [Theobroma cacao] |
| 20 |
Hb_004109_070 |
0.3670586499 |
- |
- |
hypothetical protein PRUPE_ppa010934mg [Prunus persica] |