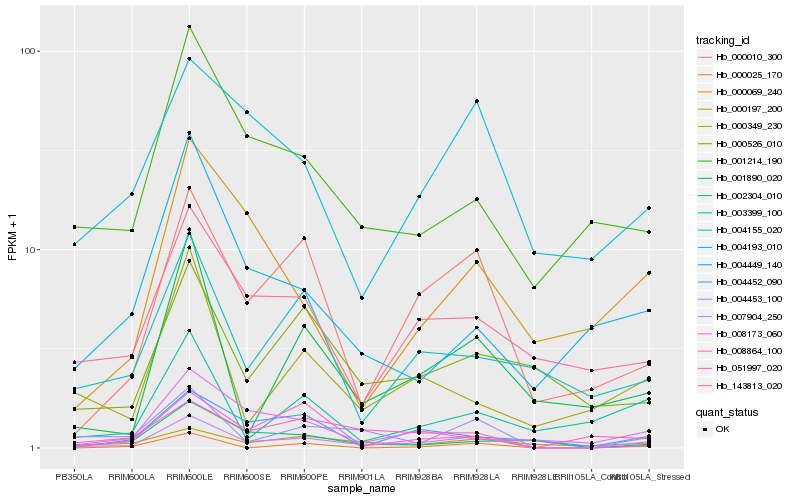
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007904_250 |
0.0 |
- |
- |
PREDICTED: vignain-like [Jatropha curcas] |
| 2 |
Hb_004453_100 |
0.191847328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000010_300 |
0.2134068646 |
transcription factor |
TF Family: TAZ |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_001214_190 |
0.244259373 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 5 |
Hb_143813_020 |
0.2516852401 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 6 |
Hb_004193_010 |
0.2545274671 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 7 |
Hb_008173_060 |
0.2562216891 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 8 |
Hb_000197_200 |
0.2565711416 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 9 |
Hb_001890_020 |
0.2575157459 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 10 |
Hb_051997_020 |
0.2577234005 |
- |
- |
PREDICTED: uncharacterized protein LOC100266863 [Vitis vinifera] |
| 11 |
Hb_000349_230 |
0.2587052084 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 12 |
Hb_002304_010 |
0.2626239336 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 13 |
Hb_004452_090 |
0.2636793411 |
- |
- |
hypothetical protein JCGZ_12231 [Jatropha curcas] |
| 14 |
Hb_004449_140 |
0.2662762437 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 15 |
Hb_004155_020 |
0.2672991704 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 16 |
Hb_000526_010 |
0.2693053885 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 17 |
Hb_000025_170 |
0.2720774707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003399_100 |
0.273419083 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 19 |
Hb_000069_240 |
0.2741205059 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3-like [Jatropha curcas] |
| 20 |
Hb_008864_100 |
0.2742125639 |
- |
- |
PREDICTED: uncharacterized protein LOC105631832 isoform X1 [Jatropha curcas] |