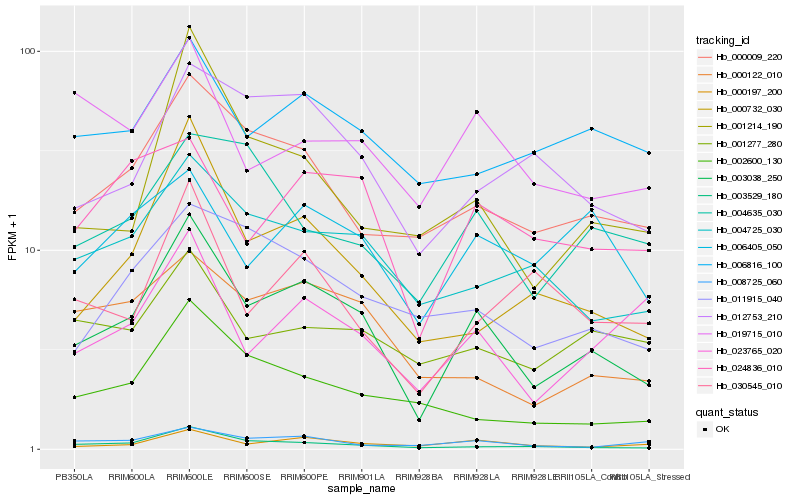
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003038_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650395 [Jatropha curcas] |
| 2 |
Hb_003529_180 |
0.1576605997 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |
| 3 |
Hb_000009_220 |
0.161747197 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_000197_200 |
0.1682334376 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 5 |
Hb_004725_030 |
0.1702910953 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X4 [Jatropha curcas] |
| 6 |
Hb_000732_030 |
0.1705303112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000122_010 |
0.1820794303 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_012753_210 |
0.1822157791 |
- |
- |
PREDICTED: inactive rhomboid protein 1-like [Jatropha curcas] |
| 9 |
Hb_011915_040 |
0.1838744736 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 10 |
Hb_024836_010 |
0.1851551634 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 11 |
Hb_002600_130 |
0.1873321421 |
- |
- |
PREDICTED: uncharacterized protein LOC105646808 [Jatropha curcas] |
| 12 |
Hb_001214_190 |
0.1876212241 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 13 |
Hb_008725_060 |
0.1906922615 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 14 |
Hb_006816_100 |
0.1913418656 |
- |
- |
hypothetical protein CISIN_1g0373181mg, partial [Citrus sinensis] |
| 15 |
Hb_023765_020 |
0.1918140716 |
- |
- |
PREDICTED: putative SNAP25 homologous protein SNAP30 [Populus euphratica] |
| 16 |
Hb_001277_280 |
0.1951305243 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 17 |
Hb_006405_050 |
0.195884345 |
- |
- |
PREDICTED: stomatal closure-related actin-binding protein 2-like [Jatropha curcas] |
| 18 |
Hb_019715_010 |
0.198502867 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 19 |
Hb_004635_030 |
0.2015011155 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 20 |
Hb_030545_010 |
0.2023226274 |
- |
- |
PREDICTED: uncharacterized protein LOC105648741 [Jatropha curcas] |