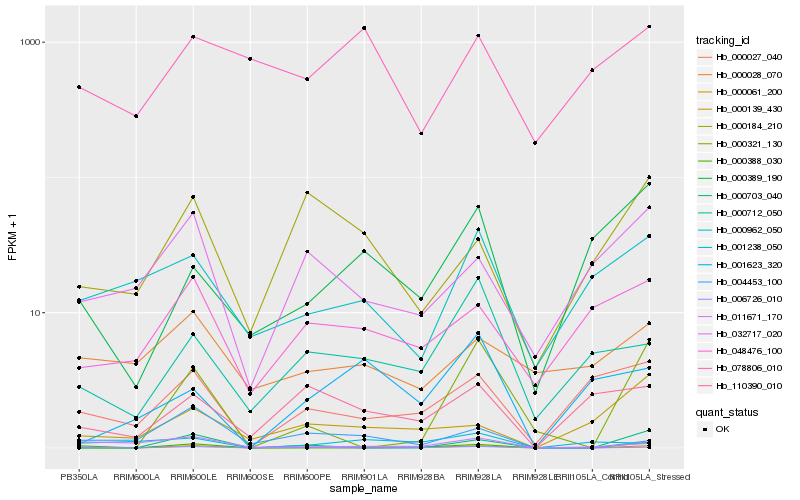
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_200 |
0.0 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 2 |
Hb_000388_030 |
0.3177272347 |
- |
- |
hypothetical protein JCGZ_13427 [Jatropha curcas] |
| 3 |
Hb_006726_010 |
0.3266610986 |
- |
- |
PREDICTED: uncharacterized protein K02A2.6-like [Nicotiana sylvestris] |
| 4 |
Hb_000027_040 |
0.3451593933 |
- |
- |
PREDICTED: uncharacterized protein LOC105636980 [Jatropha curcas] |
| 5 |
Hb_000389_190 |
0.3502445208 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A1-like [Jatropha curcas] |
| 6 |
Hb_004453_100 |
0.3507442886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000712_050 |
0.3528505849 |
- |
- |
PREDICTED: inositol oxygenase 1-like isoform X1 [Citrus sinensis] |
| 8 |
Hb_048476_100 |
0.3542135106 |
- |
- |
PREDICTED: glutathione S-transferase U9-like [Vitis vinifera] |
| 9 |
Hb_011671_170 |
0.3552362058 |
- |
- |
PREDICTED: protein DYAD [Jatropha curcas] |
| 10 |
Hb_000184_210 |
0.3552974551 |
- |
- |
RING-H2 finger protein ATL3J, putative [Ricinus communis] |
| 11 |
Hb_032717_020 |
0.3596054336 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000139_430 |
0.3663769766 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC104882266 isoform X2 [Vitis vinifera] |
| 13 |
Hb_110390_010 |
0.3711225142 |
- |
- |
hypothetical protein JCGZ_19741 [Jatropha curcas] |
| 14 |
Hb_000962_050 |
0.3750163366 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |
| 15 |
Hb_000321_130 |
0.3766579649 |
- |
- |
Alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_001238_050 |
0.3771869313 |
- |
- |
- |
| 17 |
Hb_000028_070 |
0.3777085179 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 18 |
Hb_001623_320 |
0.3791630949 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 isoform X1 [Populus euphratica] |
| 19 |
Hb_000703_040 |
0.3796006518 |
- |
- |
hypothetical protein RCOM_1177160 [Ricinus communis] |
| 20 |
Hb_078806_010 |
0.3810855012 |
- |
- |
hypothetical protein CISIN_1g031839mg [Citrus sinensis] |