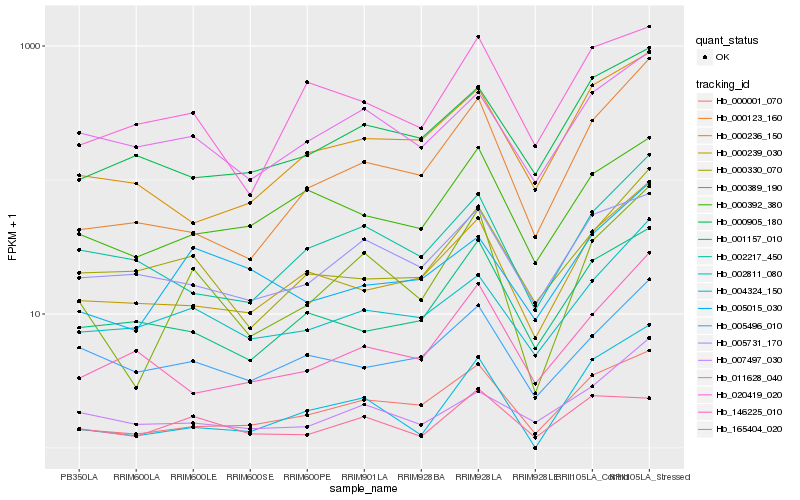
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000389_190 |
0.0 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A1-like [Jatropha curcas] |
| 2 |
Hb_000001_070 |
0.1452249306 |
- |
- |
- |
| 3 |
Hb_000123_160 |
0.1687449665 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 4 |
Hb_000239_030 |
0.1749726036 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 5 |
Hb_002217_450 |
0.1799756817 |
- |
- |
PREDICTED: probable prefoldin subunit 3 [Jatropha curcas] |
| 6 |
Hb_002811_080 |
0.1802182857 |
- |
- |
hypothetical protein POPTR_0001s08090g [Populus trichocarpa] |
| 7 |
Hb_004324_150 |
0.1805942761 |
- |
- |
- |
| 8 |
Hb_000330_070 |
0.1821716366 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 9 |
Hb_011628_040 |
0.1854469647 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 10 |
Hb_165404_020 |
0.1876328915 |
- |
- |
hypothetical protein JCGZ_17789 [Jatropha curcas] |
| 11 |
Hb_005496_010 |
0.1894580381 |
- |
- |
PREDICTED: PXMP2/4 family protein 4 [Jatropha curcas] |
| 12 |
Hb_005015_030 |
0.1915863845 |
- |
- |
PREDICTED: uncharacterized protein LOC105636546 [Jatropha curcas] |
| 13 |
Hb_000392_380 |
0.1917912731 |
- |
- |
ubiquitin-conjugating enzyme [Medicago truncatula] |
| 14 |
Hb_005731_170 |
0.1937070542 |
- |
- |
hypothetical protein PRUPE_ppa011750mg [Prunus persica] |
| 15 |
Hb_001157_010 |
0.1938762456 |
- |
- |
deoxyhypusine synthase, putative [Ricinus communis] |
| 16 |
Hb_020419_020 |
0.1969428982 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 17 |
Hb_146225_010 |
0.1982942723 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 18 |
Hb_000236_150 |
0.1983349325 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 19 |
Hb_000905_180 |
0.1986983739 |
- |
- |
60S ribosomal protein L31A [Hevea brasiliensis] |
| 20 |
Hb_007497_030 |
0.199482679 |
- |
- |
mutt domain protein, putative [Ricinus communis] |