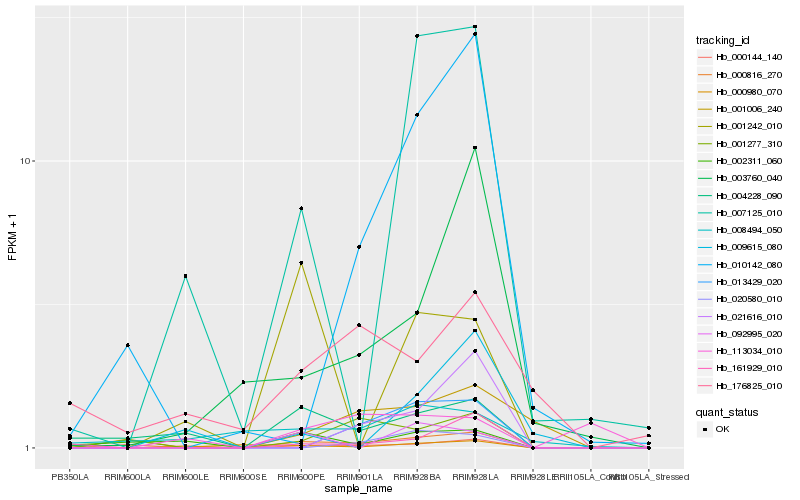
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_270 |
0.0 |
- |
- |
- |
| 2 |
Hb_004228_090 |
0.2648984411 |
- |
- |
- |
| 3 |
Hb_007125_010 |
0.2905901642 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 4 |
Hb_002311_060 |
0.3099504915 |
- |
- |
PREDICTED: protein trichome birefringence-like 4 [Jatropha curcas] |
| 5 |
Hb_161929_010 |
0.3324684467 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 6 |
Hb_003760_040 |
0.3340232806 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 7 |
Hb_001277_310 |
0.3367741731 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Phoenix dactylifera] |
| 8 |
Hb_000144_140 |
0.338721776 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 9 |
Hb_013429_020 |
0.3418466642 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 10 |
Hb_000980_070 |
0.3449816233 |
- |
- |
PREDICTED: uncharacterized protein LOC103440883 [Malus domestica] |
| 11 |
Hb_092995_020 |
0.3505940097 |
- |
- |
- |
| 12 |
Hb_113034_010 |
0.3516061397 |
- |
- |
- |
| 13 |
Hb_176825_010 |
0.3541544318 |
- |
- |
PREDICTED: uncharacterized protein LOC105630857 isoform X2 [Jatropha curcas] |
| 14 |
Hb_009615_080 |
0.3546093436 |
- |
- |
hypothetical protein JCGZ_24461 [Jatropha curcas] |
| 15 |
Hb_008494_050 |
0.3562779103 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 16 |
Hb_020580_010 |
0.3594267839 |
- |
- |
- |
| 17 |
Hb_021616_010 |
0.3610526301 |
- |
- |
- |
| 18 |
Hb_001242_010 |
0.36518061 |
- |
- |
hypothetical protein POPTR_0005s21760g [Populus trichocarpa] |
| 19 |
Hb_001006_240 |
0.3653138316 |
- |
- |
- |
| 20 |
Hb_010142_080 |
0.3660822197 |
- |
- |
PREDICTED: uncharacterized protein LOC105630660 [Jatropha curcas] |