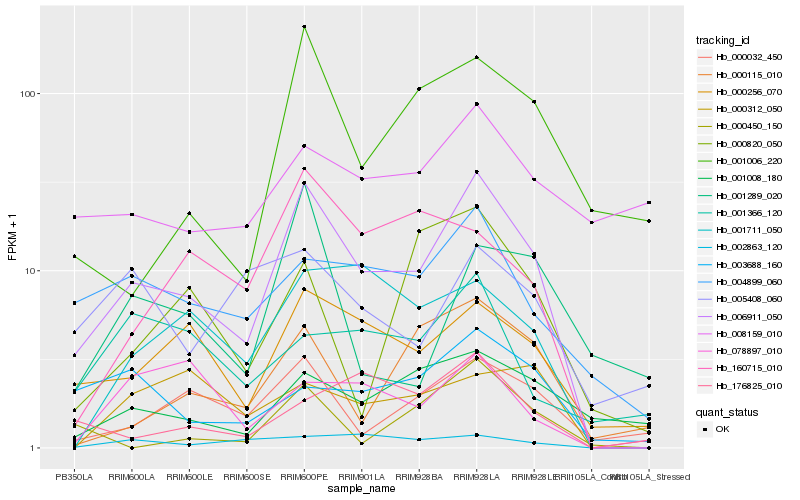
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006911_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 2 |
Hb_000256_070 |
0.2135989264 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000032_450 |
0.234712157 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 4 |
Hb_004899_060 |
0.2361397841 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 5 |
Hb_001711_050 |
0.2574252078 |
- |
- |
- |
| 6 |
Hb_001008_180 |
0.2623281885 |
- |
- |
- |
| 7 |
Hb_001006_220 |
0.2627140695 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 8 |
Hb_000115_010 |
0.2639545417 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_160715_010 |
0.2654829373 |
- |
- |
- |
| 10 |
Hb_003688_160 |
0.2655773525 |
- |
- |
Presenilin-like family protein [Populus trichocarpa] |
| 11 |
Hb_002863_120 |
0.2744917906 |
desease resistance |
Gene Name: AAA_17 |
JHL25H03.3 [Jatropha curcas] |
| 12 |
Hb_176825_010 |
0.2822762231 |
- |
- |
PREDICTED: uncharacterized protein LOC105630857 isoform X2 [Jatropha curcas] |
| 13 |
Hb_005408_060 |
0.2849393221 |
- |
- |
PREDICTED: uncharacterized protein LOC105648045 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000820_050 |
0.2865366659 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 15 |
Hb_001366_120 |
0.2874174311 |
- |
- |
unknown [Zea mays] |
| 16 |
Hb_000450_150 |
0.2876419748 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 17 |
Hb_078897_010 |
0.2883538087 |
- |
- |
hypothetical protein POPTR_0012s044902g, partial [Populus trichocarpa] |
| 18 |
Hb_001289_020 |
0.288424151 |
- |
- |
PREDICTED: uncharacterized protein LOC105635827 [Jatropha curcas] |
| 19 |
Hb_008159_010 |
0.2931235478 |
- |
- |
PREDICTED: 66 kDa stress protein [Jatropha curcas] |
| 20 |
Hb_000312_050 |
0.2952242596 |
- |
- |
transcription factor, putative [Ricinus communis] |