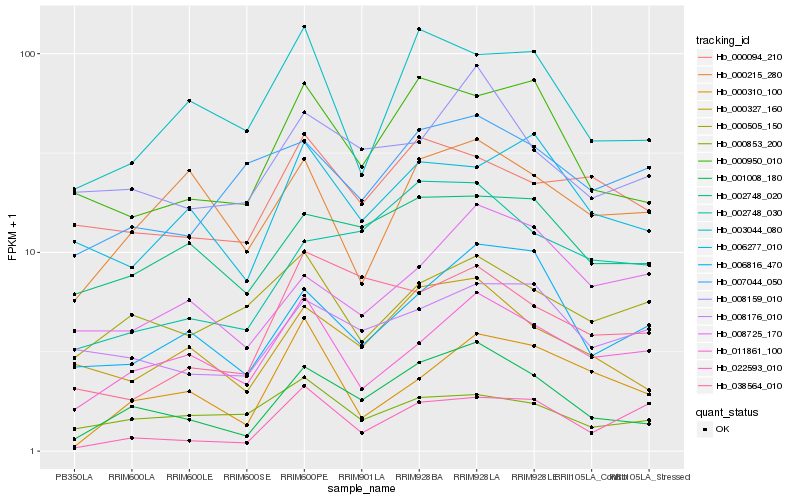
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001008_180 |
0.0 |
- |
- |
- |
| 2 |
Hb_000327_160 |
0.1444540814 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000950_010 |
0.1569690749 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 4 |
Hb_002748_030 |
0.161143301 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 5 |
Hb_006816_470 |
0.1632126147 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 6 |
Hb_008159_010 |
0.1697548153 |
- |
- |
PREDICTED: 66 kDa stress protein [Jatropha curcas] |
| 7 |
Hb_002748_020 |
0.1738719633 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 8 |
Hb_011861_100 |
0.1832314413 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_008176_010 |
0.187151543 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20090 [Jatropha curcas] |
| 10 |
Hb_003044_080 |
0.1880676375 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 11 |
Hb_038564_010 |
0.1935920642 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 12 |
Hb_006277_010 |
0.1961258488 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Gossypium raimondii] |
| 13 |
Hb_008725_170 |
0.1992905966 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Populus euphratica] |
| 14 |
Hb_000215_280 |
0.2015353586 |
- |
- |
Protein-tyrosine phosphatase mitochondrial 1, mitochondrial precursor, putative [Ricinus communis] |
| 15 |
Hb_000310_100 |
0.2041852965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000094_210 |
0.2048155278 |
- |
- |
glucose-6-phosphate isomerase, putative [Ricinus communis] |
| 17 |
Hb_000853_200 |
0.206618081 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 18 |
Hb_007044_050 |
0.209166774 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 19 |
Hb_022593_010 |
0.2097861033 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000505_150 |
0.20979836 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |