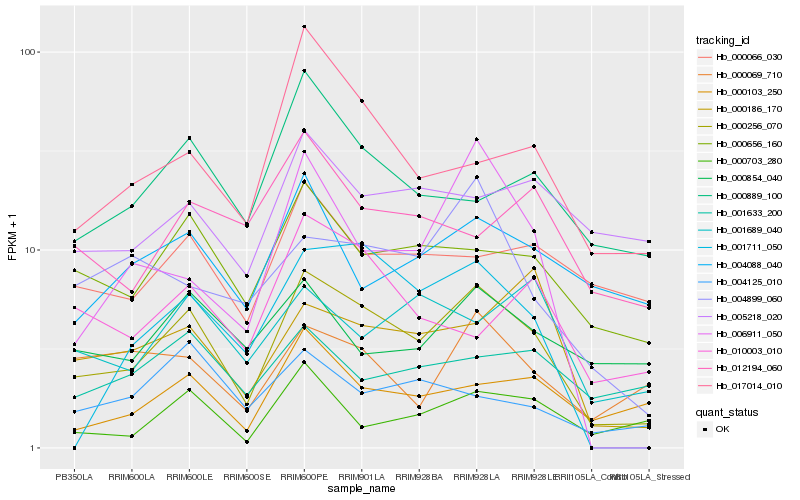
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000256_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000656_160 |
0.1710226615 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 3 |
Hb_000103_250 |
0.1976066908 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 4B isoform X3 [Jatropha curcas] |
| 4 |
Hb_000889_100 |
0.2042699456 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 5 |
Hb_017014_010 |
0.2050821444 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_004088_040 |
0.2069256853 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' iota isoform [Jatropha curcas] |
| 7 |
Hb_004899_060 |
0.2084687019 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 8 |
Hb_000186_170 |
0.209365296 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 9 |
Hb_000066_030 |
0.2100095799 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001711_050 |
0.2126744563 |
- |
- |
- |
| 11 |
Hb_000854_040 |
0.2134392458 |
- |
- |
PREDICTED: GPI mannosyltransferase 3 [Jatropha curcas] |
| 12 |
Hb_006911_050 |
0.2135989264 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 13 |
Hb_000069_710 |
0.2144321437 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |
| 14 |
Hb_010003_010 |
0.2144558994 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000703_280 |
0.2153674837 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 16 |
Hb_005218_020 |
0.2192327272 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 17 |
Hb_004125_010 |
0.2195744325 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 18 |
Hb_001689_040 |
0.2196645374 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 1-like [Jatropha curcas] |
| 19 |
Hb_012194_060 |
0.2223358114 |
- |
- |
PREDICTED: uncharacterized protein LOC105636108 [Jatropha curcas] |
| 20 |
Hb_001633_200 |
0.2226818093 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |