| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
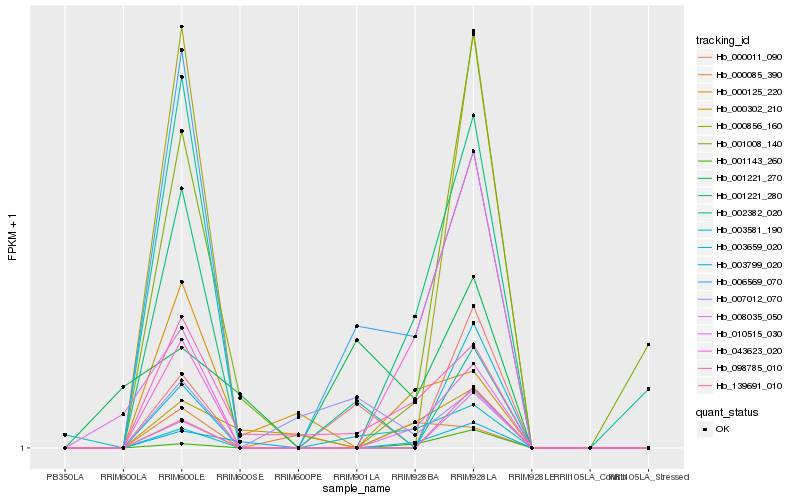
Hb_006569_070 |
0.0 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_098785_010 |
0.233480618 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 3 |
Hb_001221_280 |
0.2818275138 |
- |
- |
- |
| 4 |
Hb_000011_090 |
0.3087396858 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940-like [Jatropha curcas] |
| 5 |
Hb_007012_070 |
0.3162592514 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 6 |
Hb_003581_190 |
0.3405694991 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_21694 [Jatropha curcas] |
| 7 |
Hb_010515_030 |
0.3520971103 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 8 |
Hb_000856_160 |
0.3556056334 |
- |
- |
- |
| 9 |
Hb_000125_220 |
0.3663806804 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 13 [Jatropha curcas] |
| 10 |
Hb_003799_020 |
0.3690409951 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 11 |
Hb_002382_020 |
0.3724951277 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |
| 12 |
Hb_043623_020 |
0.374831533 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 13 |
Hb_001221_270 |
0.3927360699 |
- |
- |
- |
| 14 |
Hb_000085_390 |
0.3958535056 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_001008_140 |
0.3972264393 |
- |
- |
- |
| 16 |
Hb_003659_020 |
0.4067864602 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 17 |
Hb_001143_260 |
0.4083593373 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 18 |
Hb_008035_050 |
0.4092195254 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 19 |
Hb_139691_010 |
0.4140458357 |
- |
- |
- |
| 20 |
Hb_000302_210 |
0.414141724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |