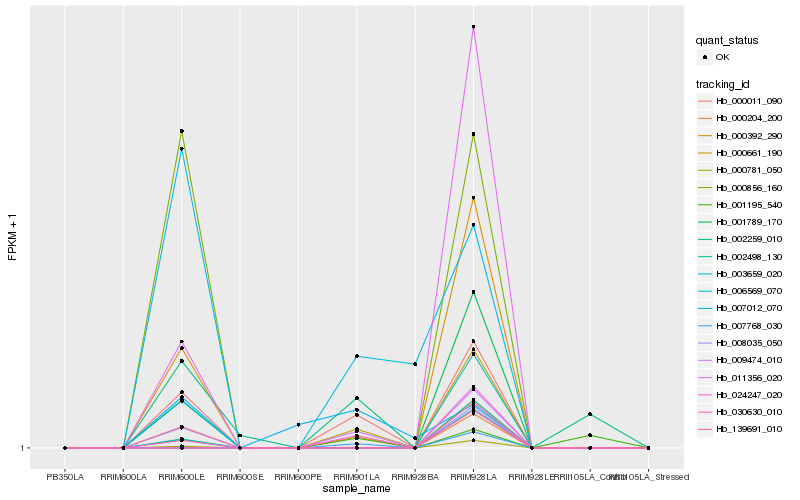
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940-like [Jatropha curcas] |
| 2 |
Hb_003659_020 |
0.2942151403 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 3 |
Hb_008035_050 |
0.2943252288 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 4 |
Hb_139691_010 |
0.2947672821 |
- |
- |
- |
| 5 |
Hb_000392_290 |
0.3014703025 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_006569_070 |
0.3087396858 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_001789_170 |
0.3131870707 |
- |
- |
hypothetical protein JCGZ_08269 [Jatropha curcas] |
| 8 |
Hb_000856_160 |
0.3256307137 |
- |
- |
- |
| 9 |
Hb_002498_130 |
0.3258635622 |
- |
- |
hypothetical protein POPTR_0004s00980g [Populus trichocarpa] |
| 10 |
Hb_000781_050 |
0.3287396223 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_011356_020 |
0.3370702459 |
- |
- |
- |
| 12 |
Hb_024247_020 |
0.3732722676 |
- |
- |
PREDICTED: uncharacterized protein LOC105634579 [Jatropha curcas] |
| 13 |
Hb_007012_070 |
0.3760719497 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 14 |
Hb_001195_540 |
0.3888229342 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 15 |
Hb_000204_200 |
0.3951626152 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 16 |
Hb_009474_010 |
0.3951682139 |
- |
- |
- |
| 17 |
Hb_030630_010 |
0.3953086366 |
- |
- |
PREDICTED: uncharacterized protein LOC105786776 [Gossypium raimondii] |
| 18 |
Hb_007768_030 |
0.3955185403 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_000661_190 |
0.4019241446 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 20 |
Hb_002259_010 |
0.4088797833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |