| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
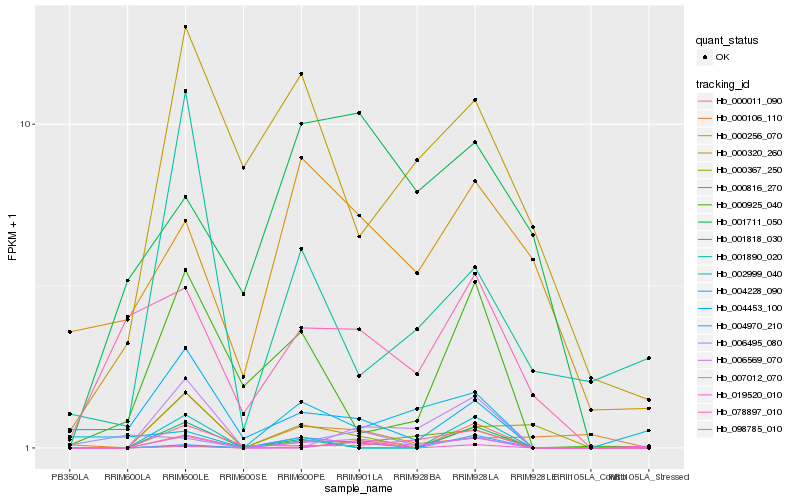
Hb_007012_070 |
0.0 |
- |
- |
PREDICTED: probable pectinesterase 53 [Jatropha curcas] |
| 2 |
Hb_006569_070 |
0.3162592514 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_098785_010 |
0.3368328155 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 4 |
Hb_004970_210 |
0.3433288859 |
- |
- |
- |
| 5 |
Hb_078897_010 |
0.3636510463 |
- |
- |
hypothetical protein POPTR_0012s044902g, partial [Populus trichocarpa] |
| 6 |
Hb_004453_100 |
0.3739407061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001711_050 |
0.3755688969 |
- |
- |
- |
| 8 |
Hb_000011_090 |
0.3760719497 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940-like [Jatropha curcas] |
| 9 |
Hb_000925_040 |
0.3804316545 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000816_270 |
0.3909699953 |
- |
- |
- |
| 11 |
Hb_000320_260 |
0.3931611149 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000367_250 |
0.3993379857 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_006495_080 |
0.4016883634 |
- |
- |
hypothetical protein PRUPE_ppb025438mg, partial [Prunus persica] |
| 14 |
Hb_000256_070 |
0.4026386854 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000106_110 |
0.4030246611 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 16 |
Hb_001818_030 |
0.4044813635 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 17 |
Hb_004228_090 |
0.4065434476 |
- |
- |
- |
| 18 |
Hb_002999_040 |
0.4068830277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001890_020 |
0.4083662516 |
- |
- |
hypothetical protein CICLE_v10025198mg [Citrus clementina] |
| 20 |
Hb_019520_010 |
0.4108322853 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |