| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
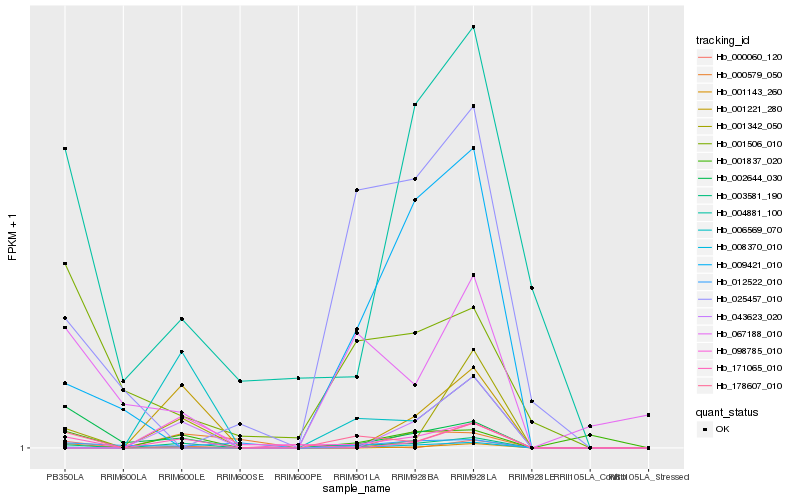
Hb_003581_190 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_21694 [Jatropha curcas] |
| 2 |
Hb_171065_010 |
0.315754425 |
- |
- |
Ribonuclease H protein [Theobroma cacao] |
| 3 |
Hb_001342_050 |
0.3332471975 |
- |
- |
- |
| 4 |
Hb_001837_020 |
0.3348374293 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X4 [Populus euphratica] |
| 5 |
Hb_008370_010 |
0.3393902584 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_006569_070 |
0.3405694991 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_004881_100 |
0.3524068357 |
- |
- |
Phosphoribosyltransferase family protein isoform 4 [Theobroma cacao] |
| 8 |
Hb_067188_010 |
0.3527147697 |
- |
- |
- |
| 9 |
Hb_002644_030 |
0.3554228979 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like, partial [Eucalyptus grandis] |
| 10 |
Hb_178607_010 |
0.3581958067 |
- |
- |
PREDICTED: uncharacterized protein LOC105789535 [Gossypium raimondii] |
| 11 |
Hb_043623_020 |
0.3596415762 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 12 |
Hb_009421_010 |
0.3693852114 |
- |
- |
- |
| 13 |
Hb_001221_280 |
0.3709768641 |
- |
- |
- |
| 14 |
Hb_000060_120 |
0.377191837 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 15 |
Hb_001143_260 |
0.3793017609 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 16 |
Hb_025457_010 |
0.38129454 |
- |
- |
- |
| 17 |
Hb_098785_010 |
0.3871729266 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 18 |
Hb_000579_050 |
0.3937975938 |
- |
- |
PREDICTED: peroxidase 9 [Jatropha curcas] |
| 19 |
Hb_012522_010 |
0.396766792 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 20 |
Hb_001506_010 |
0.3969107862 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |