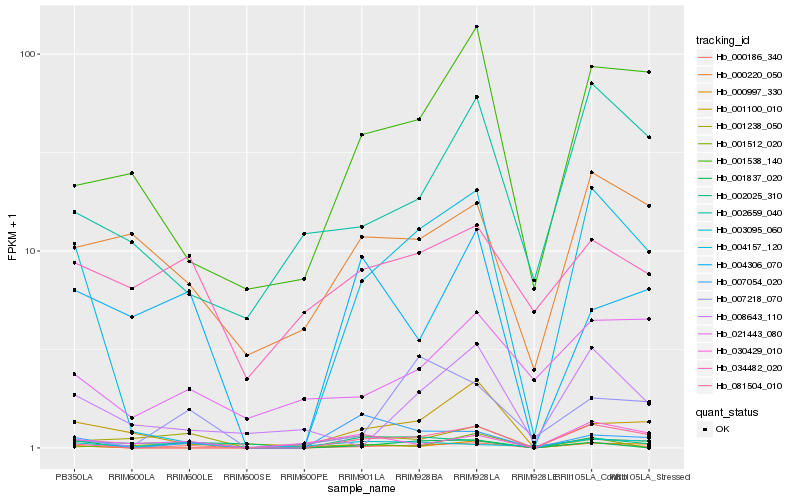
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_340 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01440g [Populus trichocarpa] |
| 2 |
Hb_001837_020 |
0.2491330422 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X4 [Populus euphratica] |
| 3 |
Hb_004157_120 |
0.2530136761 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 4 |
Hb_081504_010 |
0.2922373561 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 5 |
Hb_000997_330 |
0.3106155577 |
- |
- |
hypothetical protein POPTR_0010s08940g [Populus trichocarpa] |
| 6 |
Hb_008643_110 |
0.3157902449 |
- |
- |
BnaCnng39750D [Brassica napus] |
| 7 |
Hb_007218_070 |
0.3223369643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004306_070 |
0.3288408658 |
- |
- |
hypothetical protein 0_18789_01, partial [Pinus taeda] |
| 9 |
Hb_001538_140 |
0.3306846544 |
- |
- |
PREDICTED: PXMP2/4 family protein 4-like [Jatropha curcas] |
| 10 |
Hb_030429_010 |
0.3363623252 |
- |
- |
hypothetical protein POPTR_0001s44580g [Populus trichocarpa] |
| 11 |
Hb_001100_010 |
0.3369478279 |
- |
- |
F-box/FBD/LRR-repeat protein [Medicago truncatula] |
| 12 |
Hb_002659_040 |
0.3376244815 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_001238_050 |
0.3384655861 |
- |
- |
- |
| 14 |
Hb_000220_050 |
0.3406053446 |
transcription factor |
TF Family: Orphans |
PREDICTED: putative zinc finger protein At1g68190 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002025_310 |
0.3420377669 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_003095_060 |
0.3439641294 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 17 |
Hb_034482_020 |
0.344525031 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_007054_020 |
0.3445532459 |
- |
- |
- |
| 19 |
Hb_001512_020 |
0.3469022412 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 20 |
Hb_021443_080 |
0.3471216488 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |