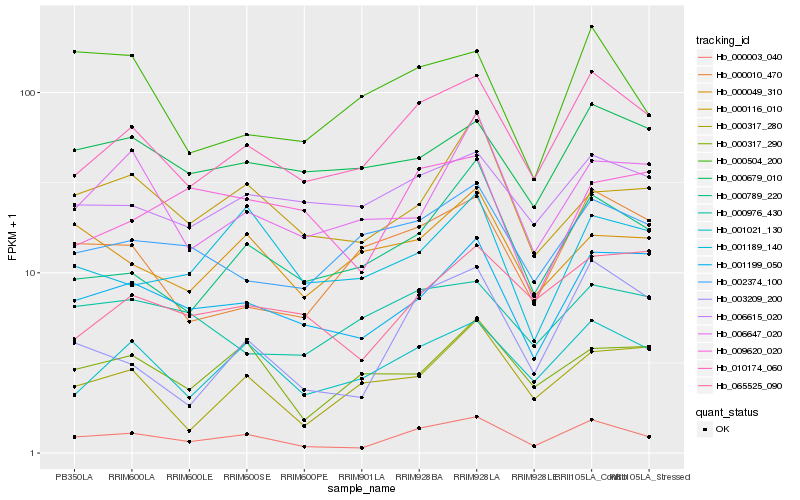
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 2 |
Hb_010174_060 |
0.1125390624 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 3 |
Hb_001021_130 |
0.1371998475 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 4 |
Hb_001199_050 |
0.1376220276 |
- |
- |
PREDICTED: DNA-3-methyladenine glycosylase-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000789_220 |
0.1517467253 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 6 |
Hb_003209_200 |
0.1537857953 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1621230 [Ricinus communis] |
| 7 |
Hb_000116_010 |
0.1553150104 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_006615_020 |
0.1672855825 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM2 [Jatropha curcas] |
| 9 |
Hb_002374_100 |
0.1674709968 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 10 |
Hb_000317_290 |
0.1683377038 |
- |
- |
PREDICTED: VQ motif-containing protein 31 [Jatropha curcas] |
| 11 |
Hb_001189_140 |
0.1683439346 |
- |
- |
- |
| 12 |
Hb_000679_010 |
0.1700071593 |
- |
- |
PREDICTED: uncharacterized protein LOC105637663 [Jatropha curcas] |
| 13 |
Hb_000049_310 |
0.1728807223 |
- |
- |
Myosin-J heavy chain, partial [Glycine soja] |
| 14 |
Hb_006647_020 |
0.1738010697 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 15 |
Hb_000010_470 |
0.1764893635 |
- |
- |
Galactose oxidase/kelch repeat superfamily protein isoform 2 [Theobroma cacao] |
| 16 |
Hb_000504_200 |
0.1787040323 |
- |
- |
hypothetical protein JCGZ_10253 [Jatropha curcas] |
| 17 |
Hb_065525_090 |
0.1797092255 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 18 |
Hb_009620_020 |
0.1812013537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000976_430 |
0.1814981459 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJD5 [Jatropha curcas] |
| 20 |
Hb_000317_280 |
0.1819957031 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At5g08490 [Jatropha curcas] |