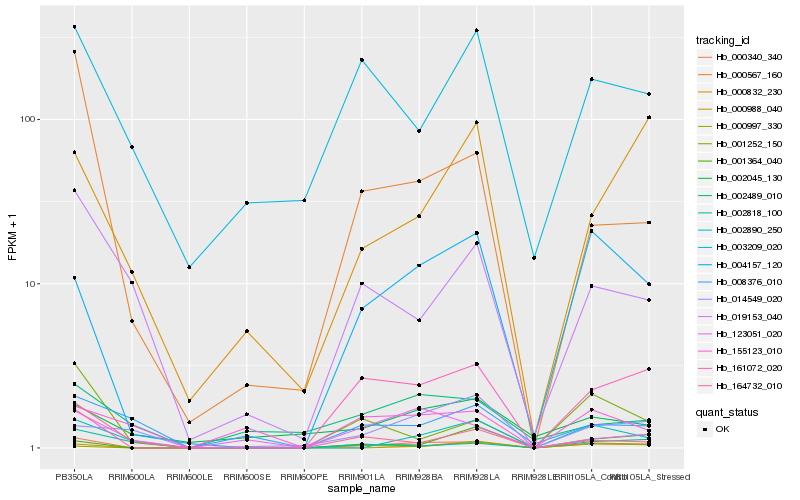
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_340 |
0.0 |
- |
- |
hypothetical protein POPTR_0014s13260g [Populus trichocarpa] |
| 2 |
Hb_004157_120 |
0.2316062971 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 3 |
Hb_000567_160 |
0.260799572 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Jatropha curcas] |
| 4 |
Hb_123051_020 |
0.2857383512 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 5 |
Hb_001252_150 |
0.2857768912 |
- |
- |
- |
| 6 |
Hb_002890_250 |
0.2877165219 |
- |
- |
PREDICTED: uncharacterized protein LOC102611387 isoform X2 [Citrus sinensis] |
| 7 |
Hb_019153_040 |
0.2940133622 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_164732_010 |
0.3010428364 |
- |
- |
- |
| 9 |
Hb_003209_020 |
0.3020277728 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 10 [Jatropha curcas] |
| 10 |
Hb_000988_040 |
0.3036287918 |
- |
- |
- |
| 11 |
Hb_161072_020 |
0.3075406995 |
- |
- |
- |
| 12 |
Hb_000997_330 |
0.3169230549 |
- |
- |
hypothetical protein POPTR_0010s08940g [Populus trichocarpa] |
| 13 |
Hb_002489_010 |
0.3208911778 |
- |
- |
hypothetical protein JCGZ_17199 [Jatropha curcas] |
| 14 |
Hb_000832_230 |
0.3227797219 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_18434 [Jatropha curcas] |
| 15 |
Hb_001364_040 |
0.3274261428 |
- |
- |
- |
| 16 |
Hb_008376_010 |
0.3277891781 |
- |
- |
- |
| 17 |
Hb_014549_020 |
0.3280308573 |
- |
- |
hypothetical protein POPTR_0013s12350g [Populus trichocarpa] |
| 18 |
Hb_002045_130 |
0.3285391889 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 19 |
Hb_155123_010 |
0.3309399142 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 20 |
Hb_002818_100 |
0.3343504458 |
- |
- |
hypothetical protein JCGZ_14122 [Jatropha curcas] |