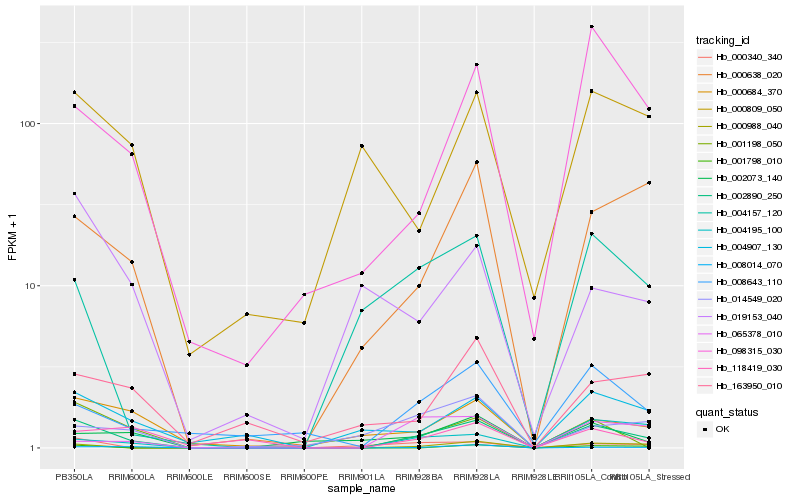
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102611387 isoform X2 [Citrus sinensis] |
| 2 |
Hb_000988_040 |
0.2295869761 |
- |
- |
- |
| 3 |
Hb_001198_050 |
0.2460817197 |
- |
- |
hypothetical protein POPTR_0004s22990g [Populus trichocarpa] |
| 4 |
Hb_000638_020 |
0.2536459314 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 9-like [Jatropha curcas] |
| 5 |
Hb_008014_070 |
0.258974027 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_000684_370 |
0.2660336477 |
- |
- |
- |
| 7 |
Hb_004907_130 |
0.2692751681 |
- |
- |
- |
| 8 |
Hb_098315_030 |
0.2757872224 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 9 |
Hb_008643_110 |
0.2781800154 |
- |
- |
BnaCnng39750D [Brassica napus] |
| 10 |
Hb_000340_340 |
0.2877165219 |
- |
- |
hypothetical protein POPTR_0014s13260g [Populus trichocarpa] |
| 11 |
Hb_163950_010 |
0.294480245 |
- |
- |
- |
| 12 |
Hb_004157_120 |
0.2987697324 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Jatropha curcas] |
| 13 |
Hb_014549_020 |
0.300693103 |
- |
- |
hypothetical protein POPTR_0013s12350g [Populus trichocarpa] |
| 14 |
Hb_002073_140 |
0.3038440901 |
- |
- |
- |
| 15 |
Hb_118419_030 |
0.3052868018 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2-like [Jatropha curcas] |
| 16 |
Hb_000809_050 |
0.3079600015 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639663 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004195_100 |
0.3079678287 |
- |
- |
PREDICTED: uncharacterized protein LOC104106427 [Nicotiana tomentosiformis] |
| 18 |
Hb_019153_040 |
0.3091879805 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 19 |
Hb_001798_010 |
0.3104434756 |
transcription factor |
TF Family: NAC |
NAC domain containing protein 2-like protein, partial [Helianthus annuus] |
| 20 |
Hb_065378_010 |
0.3146867827 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |