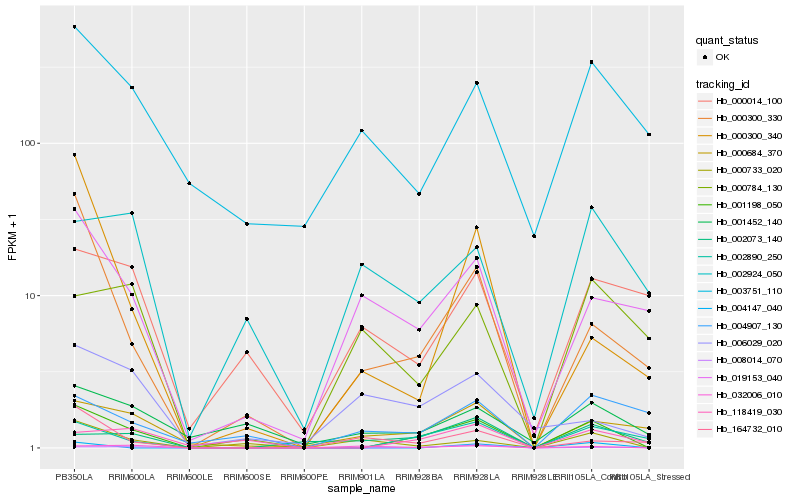
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001198_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s22990g [Populus trichocarpa] |
| 2 |
Hb_002890_250 |
0.2460817197 |
- |
- |
PREDICTED: uncharacterized protein LOC102611387 isoform X2 [Citrus sinensis] |
| 3 |
Hb_000300_330 |
0.3090325266 |
- |
- |
- |
| 4 |
Hb_000684_370 |
0.313055508 |
- |
- |
- |
| 5 |
Hb_118419_030 |
0.3321050573 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2-like [Jatropha curcas] |
| 6 |
Hb_000733_020 |
0.3334736977 |
- |
- |
- |
| 7 |
Hb_019153_040 |
0.334827653 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_008014_070 |
0.3372289641 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_004147_040 |
0.3447173203 |
- |
- |
hypothetical protein JCGZ_15918 [Jatropha curcas] |
| 10 |
Hb_003751_110 |
0.3452358861 |
- |
- |
unknown [Lotus japonicus] |
| 11 |
Hb_001452_140 |
0.3485392435 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 12 |
Hb_164732_010 |
0.3575361958 |
- |
- |
- |
| 13 |
Hb_032006_010 |
0.3579808325 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 14 |
Hb_000300_340 |
0.3605913215 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_002073_140 |
0.3623727715 |
- |
- |
- |
| 16 |
Hb_006029_020 |
0.3679665911 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1, partial [Vitis vinifera] |
| 17 |
Hb_004907_130 |
0.3690070769 |
- |
- |
- |
| 18 |
Hb_000784_130 |
0.3713391002 |
- |
- |
hypothetical protein JCGZ_22813 [Jatropha curcas] |
| 19 |
Hb_002924_050 |
0.3745382182 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 20 |
Hb_000014_100 |
0.3747619178 |
- |
- |
- |